6SYE
 
 | |
6SYM
 
 | |
2WK0
 
 | | Crystal structure of the class A beta-lactamase BS3 inhibited by 6- beta-iodopenicillanate. | | 分子名称: | (3S)-2,2-dimethyl-3,4-dihydro-2H-1,4-thiazine-3,6-dicarboxylic acid, BETA-LACTAMASE, CHLORIDE ION, ... | | 著者 | Sauvage, E, Zervosen, A, Dive, G, Herman, R, Kerff, F, Amoroso, A, Fonze, E, Pratt, R.F, Luxen, A, Charlier, P. | | 登録日 | 2009-06-03 | | 公開日 | 2009-12-01 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural Basis of the Inhibition of Class a Beta-Lactamases and Penicillin-Binding Proteins by 6-Beta-Iodopenicillanate.
J.Am.Chem.Soc., 131, 2009
|
|
2WJ8
 
 | | Respiratory Syncitial Virus RiboNucleoProtein | | 分子名称: | BORATE ION, NUCLEOPROTEIN, RNA (5'-R(*CP*CP*CP*CP*CP*C)-3') | | 著者 | Tawar, R.G, Duquerroy, S, Vonrhein, C, Varela, P.F, Damier-Piolle, L, Castagne, N, MacLellan, K, Bedouelle, H, Bricogne, G, Bhella, D, Eleouet, J, Rey, F.A. | | 登録日 | 2009-05-25 | | 公開日 | 2009-12-08 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (3.29 Å) | | 主引用文献 | Crystal Structure of a Nucleocapsid-Like Nucleoprotein-RNA Complex of Respiratory Syncytial Virus
Science, 326, 2009
|
|
4ZE4
 
 | | Structure of Gan1D, a putative 6-phospho-beta-galactosidase from Geobacillus stearothermophilus | | 分子名称: | GLYCEROL, IMIDAZOLE, Putative 6-phospho-beta-galactobiosidase | | 著者 | Lansky, S, Zehavi, A, Dvir, H, Shoham, Y, Shoham, G. | | 登録日 | 2015-04-20 | | 公開日 | 2016-06-29 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Structure of Gan1D, a putative 6-phospho-beta-galactosidase from Geobacillus stearothermophilus
To Be Published
|
|
2O4M
 
 | | Structure of Phosphotriesterase mutant I106G/F132G/H257Y | | 分子名称: | ACETIC ACID, CACODYLATE ION, GLYCEROL, ... | | 著者 | Kim, J, Ramagopal, U.A, Tsai, P, Raushel, F.M, Almo, S.C. | | 登録日 | 2006-12-04 | | 公開日 | 2007-12-18 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Structure of Phosphotriesterase mutant I106G/F132G/H257Y
To be Published
|
|
2JJT
 
 | | Structure of human CD47 in complex with human signal regulatory protein (SIRP) alpha | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LEUKOCYTE SURFACE ANTIGEN CD47, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE SUBSTRATE 1 | | 著者 | Hatherley, D, Graham, S.C, Turner, J, Harlos, K, Stuart, D.I, Barclay, A.N. | | 登録日 | 2008-04-22 | | 公開日 | 2008-08-05 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Paired receptor specificity explained by structures of signal regulatory proteins alone and complexed with CD47.
Mol. Cell, 31, 2008
|
|
4ZF8
 
 | | Cytochrome P450 pentamutant from BM3 with bound Metyrapone | | 分子名称: | Bifunctional P-450/NADPH-P450 reductase, METYRAPONE, NICKEL (II) ION, ... | | 著者 | Rogers, W.E, Othman, T, Heidary, D.K, Huxford, T. | | 登録日 | 2015-04-21 | | 公開日 | 2016-07-13 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.766 Å) | | 主引用文献 | Effect of Mutation and Substrate Binding on the Stability of Cytochrome P450BM3 Variants.
Biochemistry, 55, 2016
|
|
6TRS
 
 | | Crystal structure of TFIIH subunit p52 in complex with p8 | | 分子名称: | RNA polymerase II transcription factor B subunit 2, Uncharacterized protein | | 著者 | Koelmel, W, Kuper, J, Schoenwetter, E, Kisker, C. | | 登録日 | 2019-12-19 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | How to limit the speed of a motor: the intricate regulation of the XPB ATPase and translocase in TFIIH.
Nucleic Acids Res., 48, 2020
|
|
4ZOE
 
 | | Crystal Structure of beta-glucosidase from Listeria innocua | | 分子名称: | GLYCEROL, Lin1840 protein, MAGNESIUM ION | | 著者 | Nakajima, M, Yoshida, R, Miyanaga, A, Abe, K, Takahashi, Y, Sugimoto, N, Toyoizumi, H, Nakai, H, Kitaoka, M, Taguchi, H. | | 登録日 | 2015-05-06 | | 公開日 | 2016-05-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Functional and Structural Analysis of a beta-Glucosidase Involved in beta-1,2-Glucan Metabolism in Listeria innocua
Plos One, 11, 2016
|
|
2O7P
 
 | | The crystal structure of RibD from Escherichia coli in complex with the oxidised NADP+ cofactor in the active site of the reductase domain | | 分子名称: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Riboflavin biosynthesis protein ribD | | 著者 | Moche, M, Stenmark, P, Gurmu, D, Nordlund, P, Structural Proteomics in Europe (SPINE) | | 登録日 | 2006-12-11 | | 公開日 | 2007-02-13 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The crystal structure of the bifunctional deaminase/reductase RibD of the riboflavin biosynthetic pathway in Escherichia coli: implications for the reductive mechanism.
J.Mol.Biol., 373, 2007
|
|
2OB5
 
 | | Crystal structure of protein Atu2016, putative sugar binding protein | | 分子名称: | Hypothetical protein Atu2016 | | 著者 | Chang, C, Xu, X, Gu, J, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2006-12-18 | | 公開日 | 2007-01-16 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structure of protein Atu2016, putative sugar binding protein
To be Published
|
|
4ZHS
 
 | |
2OBC
 
 | | The crystal structure of RibD from Escherichia coli in complex with a substrate analogue, ribose 5-phosphate (beta form), bound to the active site of the reductase domain | | 分子名称: | 5-O-phosphono-beta-D-ribofuranose, Riboflavin biosynthesis protein ribD | | 著者 | Moche, M, Stenmark, P, Gurmu, D, Nordlund, P, Structural Proteomics in Europe (SPINE) | | 登録日 | 2006-12-18 | | 公開日 | 2007-02-13 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The crystal structure of the bifunctional deaminase/reductase RibD of the riboflavin biosynthetic pathway in Escherichia coli: implications for the reductive mechanism.
J.Mol.Biol., 373, 2007
|
|
6TFK
 
 | | Vip3Aa toxin structure | | 分子名称: | MAGNESIUM ION, Vegetative insecticidal protein | | 著者 | Nunez-Ramirez, R, Huesa, J, Bel, Y, Ferre, J, Casino, P, Arias-Palomo, E. | | 登録日 | 2019-11-14 | | 公開日 | 2020-08-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Molecular architecture and activation of the insecticidal protein Vip3Aa from Bacillus thuringiensis.
Nat Commun, 11, 2020
|
|
6TTE
 
 | | Beta-galactosidase in complex with PETG | | 分子名称: | 2-phenylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, MAGNESIUM ION | | 著者 | Saur, M, Hartshorn, M.J, Dong, J, Reeks, J, Bunkoczi, G, Jhoti, H, Williams, P.A. | | 登録日 | 2019-12-27 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (2.2 Å) | | 主引用文献 | Fragment-based drug discovery using cryo-EM.
Drug Discov Today, 25, 2020
|
|
6JNO
 
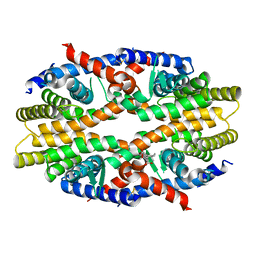 | | RXRa structure complexed with CU-6PMN | | 分子名称: | 7-oxidanyl-2-oxidanylidene-6-(3,5,5,8,8-pentamethyl-6,7-dihydronaphthalen-2-yl)chromene-3-carboxylic acid, Retinoic acid receptor RXR-alpha | | 著者 | Kawasaki, M, Nakano, S, Motoyama, T, Yamada, S, Watanabe, M, Takamura, Y, Fujihara, M, Tokiwa, H, Kakuta, H, Ito, S. | | 登録日 | 2019-03-17 | | 公開日 | 2019-11-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Competitive Binding Assay with an Umbelliferone-Based Fluorescent Rexinoid for Retinoid X Receptor Ligand Screening.
J.Med.Chem., 62, 2019
|
|
2O9R
 
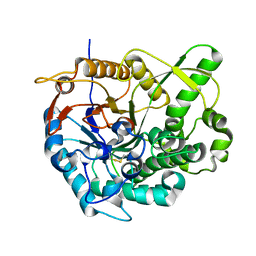 | | beta-glucosidase B complexed with thiocellobiose | | 分子名称: | Beta-glucosidase B, beta-D-glucopyranose-(1-4)-4-thio-beta-D-glucopyranose | | 著者 | Isorna, P, Polaina, J, Sanz-Aparicio, J. | | 登録日 | 2006-12-14 | | 公開日 | 2007-10-02 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal Structures of Paenibacillus polymyxa beta-Glucosidase B Complexes Reveal the Molecular Basis of Substrate Specificity and Give New Insights into the Catalytic Machinery of Family I Glycosidases
J.Mol.Biol., 371, 2007
|
|
5XYL
 
 | |
4ZMG
 
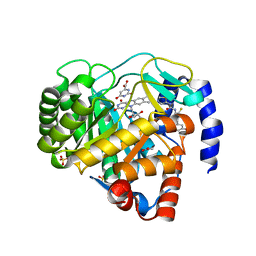 | | Crystal structure of Human Dihydroorotate Dehydrogenase (DHODH) with DH03A338 | | 分子名称: | 1-(3,5-difluoro-3'-methoxybiphenyl-4-yl)-3-(1,3-thiazol-5-yl)urea, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | 著者 | Ren, X.L, Zhu, J.S, Zhu, L.L, Li, H.L. | | 登録日 | 2015-05-04 | | 公開日 | 2016-05-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of Human Dihydroorotate Dehydrogenase (DHODH) with DH03A338
To Be Published
|
|
6ZO0
 
 | | 2.23 A resolution 3,4-dimethylcatechol (3,4-dimethylbenzene-1,2-diol) inhibited Sporosarcina pasteurii urease | | 分子名称: | 1,2-ETHANEDIOL, HYDROXIDE ION, NICKEL (II) ION, ... | | 著者 | Mazzei, L, Cianci, M, Musiani, F, Ciurli, S. | | 登録日 | 2020-07-07 | | 公開日 | 2020-12-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | Inhibition of Urease, a Ni-Enzyme: The Reactivity of a Key Thiol With Mono- and Di-Substituted Catechols Elucidated by Kinetic, Structural, and Theoretical Studies.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7ADC
 
 | | Transcription termination intermediate complex 3 delta NusG | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase subunit alpha, ... | | 著者 | Said, N, Hilal, T, Loll, B, Wahl, C.M. | | 登録日 | 2020-09-14 | | 公開日 | 2020-11-25 | | 最終更新日 | 2021-02-03 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase rho.
Science, 371, 2021
|
|
6T2V
 
 | | Cryo-EM structure of the RecBCD in complex with Chi-plus2 substrate | | 分子名称: | DNA (Chi-plus2), RecBCD enzyme subunit RecB, RecBCD enzyme subunit RecC, ... | | 著者 | Cheng, K, Wilkinson, M, Wigley, D.B. | | 登録日 | 2019-10-09 | | 公開日 | 2020-01-01 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | A conformational switch in response to Chi converts RecBCD from phage destruction to DNA repair.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4ZO9
 
 | | Crystal Structure of mutant (D270A) beta-glucosidase from Listeria innocua in complex with laminaribiose | | 分子名称: | GLYCEROL, Lin1840 protein, MAGNESIUM ION, ... | | 著者 | Nakajima, M, Yoshida, R, Miyanaga, A, Abe, K, Takahashi, Y, Sugimoto, N, Toyoizumi, H, Nakai, H, Kitaoka, M, Taguchi, H. | | 登録日 | 2015-05-06 | | 公開日 | 2016-05-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Functional and Structural Analysis of a beta-Glucosidase Involved in beta-1,2-Glucan Metabolism in Listeria innocua
Plos One, 11, 2016
|
|
6ZNY
 
 | | 1.50 A resolution 3-methylcatechol (3-methylbenzene-1,2-diol) inhibited Sporosarcina pasteurii urease | | 分子名称: | 1,2-ETHANEDIOL, HYDROXIDE ION, NICKEL (II) ION, ... | | 著者 | Mazzei, L, Cianci, M, Musiani, F, Ciurli, S. | | 登録日 | 2020-07-07 | | 公開日 | 2020-12-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Inhibition of Urease, a Ni-Enzyme: The Reactivity of a Key Thiol With Mono- and Di-Substituted Catechols Elucidated by Kinetic, Structural, and Theoretical Studies.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
