6H1E
 
 | | Crystal structure of C21orf127-TRMT112 in complex with SAH and H4 peptide | | 分子名称: | HemK methyltransferase family member 2, Histone H4 peptide, Multifunctional methyltransferase subunit TRM112-like protein, ... | | 著者 | Wang, S, Hermann, B, Metzger, E, Peng, L, Einsle, O, Schuele, R. | | 登録日 | 2018-07-11 | | 公開日 | 2019-05-22 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | KMT9 monomethylates histone H4 lysine 12 and controls proliferation of prostate cancer cells.
Nat.Struct.Mol.Biol., 26, 2019
|
|
4H4M
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with (E)-3-(3-chloro-5-(4-chloro-2-(2-(2,4-dioxo-3,4- dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)phenyl)acrylonitrile (JLJ494), a Non-nucleoside Inhibitor | | 分子名称: | (2E)-3-(3-chloro-5-{4-chloro-2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}phenyl)prop-2-enenitrile, 1,2-ETHANEDIOL, Reverse transcriptase/ribonuclease H, ... | | 著者 | Frey, K.M, Anderson, K.S. | | 登録日 | 2012-09-17 | | 公開日 | 2012-12-05 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Crystal Structures of HIV-1 Reverse Transcriptase with Picomolar Inhibitors Reveal Key Interactions for Drug Design.
J.Am.Chem.Soc., 134, 2012
|
|
1B76
 
 | |
4ERY
 
 | | X-ray structure of WDR5-MLL3 Win motif peptide binary complex | | 分子名称: | Histone-lysine N-methyltransferase MLL3, WD repeat-containing protein 5 | | 著者 | Dharmarajan, V, Lee, J.-H, Patel, A, Skalnik, D.G, Cosgrove, M.S. | | 登録日 | 2012-04-21 | | 公開日 | 2012-05-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases.
J.Biol.Chem., 287, 2012
|
|
4CFG
 
 | |
4Q0Q
 
 | | Crystal structure of Acinetobacter sp. DL28 L-ribose isomerase in complex with L-ribulose | | 分子名称: | COBALT (II) ION, COBALT HEXAMMINE(III), L-Ribose isomerase, ... | | 著者 | Yoshida, H, Yoshihara, A, Teraoka, M, Izumori, K, Kamitori, S. | | 登録日 | 2014-04-02 | | 公開日 | 2014-05-28 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | X-ray structure of a novel L-ribose isomerase acting on a non-natural sugar L-ribose as its ideal substrate.
Febs J., 281, 2014
|
|
4Q0U
 
 | | Crystal structure of Acinetobacter sp. DL28 L-ribose isomerase mutant E204Q in complex with L-ribose | | 分子名称: | COBALT (II) ION, COBALT HEXAMMINE(III), L-Ribose isomerase, ... | | 著者 | Yoshida, H, Yoshihara, A, Teraoka, M, Izumori, K, Kamitori, S. | | 登録日 | 2014-04-02 | | 公開日 | 2014-05-28 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | X-ray structure of a novel L-ribose isomerase acting on a non-natural sugar L-ribose as its ideal substrate.
Febs J., 281, 2014
|
|
4PXA
 
 | | DEAD-box RNA helicase DDX3X Cancer-associated mutant D354V | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase DDX3X, PHOSPHATE ION | | 著者 | Epling, L.B, Grace, C.R, Lowe, B.R, Partridge, J.F, Enemark, E.J. | | 登録日 | 2014-03-22 | | 公開日 | 2015-03-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Cancer-Associated Mutants of RNA Helicase DDX3X Are Defective in RNA-Stimulated ATP Hydrolysis.
J.Mol.Biol., 427, 2015
|
|
4BTE
 
 | | DJ-1 Cu(I) complex | | 分子名称: | COPPER (I) ION, PROTEIN DJ-1 | | 著者 | Puno, M.R.A, Odell, M, Moody, P.C.E. | | 登録日 | 2013-06-14 | | 公開日 | 2013-11-06 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Structure of Cu(I)-Bound Dj-1 Reveals a Biscysteinate Metal Binding Site at the Homodimer Interface: Insights Into Mutational Inactivation of Dj-1 in Parkinsonism.
J.Am.Chem.Soc., 135, 2013
|
|
4MR1
 
 | |
3EMH
 
 | | Structural basis of WDR5-MLL interaction | | 分子名称: | Mixed-lineage leukemia protein 1, SULFATE ION, WD repeat-containing protein 5 | | 著者 | Song, J.J, Kingston, R.E. | | 登録日 | 2008-09-24 | | 公開日 | 2008-10-07 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.37 Å) | | 主引用文献 | WDR5 Interacts with Mixed Lineage Leukemia (MLL) Protein via the Histone H3-binding Pocket.
J.Biol.Chem., 283, 2008
|
|
3DOK
 
 | | Crystal structure of K103N mutant HIV-1 reverse transcriptase in complex with GW678248. | | 分子名称: | 2-{4-chloro-2-[(3-chloro-5-cyanophenyl)carbonyl]phenoxy}-N-(2-methyl-4-sulfamoylphenyl)acetamide, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | 著者 | Chamberlain, P.P, Ren, J, Stammers, D.K. | | 登録日 | 2008-07-04 | | 公開日 | 2008-08-12 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural basis for the improved drug resistance profile of new generation benzophenone non-nucleoside HIV-1 reverse transcriptase inhibitors.
J.Med.Chem., 51, 2008
|
|
4B7K
 
 | | FACTOR INHIBITING HIF-1 ALPHA IN COMPLEX WITH CONSENSUS ANKYRIN REPEAT DOMAIN-SER PEPTIDE (20-MER) | | 分子名称: | CONSENSUS ANKYRIN REPEAT DOMAIN-SER, HYPOXIA-INDUCIBLE FACTOR 1-ALPHA INHIBITOR, N-OXALYLGLYCINE, ... | | 著者 | Chowdhury, R, Ge, W, Schofield, C.J. | | 登録日 | 2012-08-20 | | 公開日 | 2013-01-23 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Substrate selectivity analyses of factor inhibiting hypoxia-inducible factor.
Angew. Chem. Int. Ed. Engl., 52, 2013
|
|
4BIO
 
 | |
3DMJ
 
 | | CRYSTAL STRUCTURE OF HIV-1 V106A and Y181C MUTANT REVERSE TRANSCRIPTASE IN COMPLEX WITH GW564511. | | 分子名称: | N-{4-[amino(dihydroxy)-lambda~4~-sulfanyl]-2-methylphenyl}-2-(4-chloro-2-{[3-fluoro-5-(trifluoromethyl)phenyl]carbonyl}phenoxy)acetamide, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | 著者 | Ren, J, Chamberlain, P.P, Stammers, D.K. | | 登録日 | 2008-07-01 | | 公開日 | 2008-08-12 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis for the improved drug resistance profile of new generation benzophenone non-nucleoside HIV-1 reverse transcriptase inhibitors.
J.Med.Chem., 51, 2008
|
|
3DRP
 
 | | HIV reverse transcriptase in complex with inhibitor R8e | | 分子名称: | 3-{5-[(6-amino-1H-pyrazolo[3,4-b]pyridin-3-yl)methoxy]-2-chlorophenoxy}-5-chlorobenzonitrile, Reverse transcriptase/ribonuclease H, p51 RT | | 著者 | Yan, Y, Prasad, S. | | 登録日 | 2008-07-11 | | 公開日 | 2008-10-14 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Discovery of 3-{5-[(6-Amino-1H-pyrazolo[3,4-b]pyridine-3-yl)methoxy]-2-chlorophenoxy}-5-chlorobenzonitrile (MK-4965): A Potent, Orally Bioavailable HIV-1 Non-Nucleoside Reverse Transcriptase Inhibitor with Improved Potency against Key Mutant Viruses.
J.Med.Chem., 51, 2008
|
|
3E01
 
 | |
3DLE
 
 | |
3DM2
 
 | | Crystal structure of HIV-1 K103N mutant reverse transcriptase in complex with GW564511. | | 分子名称: | N-{4-[amino(dihydroxy)-lambda~4~-sulfanyl]-2-methylphenyl}-2-(4-chloro-2-{[3-fluoro-5-(trifluoromethyl)phenyl]carbonyl}phenoxy)acetamide, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | 著者 | Ren, J, Chamberlain, P.P, Stammers, D.K. | | 登録日 | 2008-06-30 | | 公開日 | 2008-08-12 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural basis for the improved drug resistance profile of new generation benzophenone non-nucleoside HIV-1 reverse transcriptase inhibitors.
J.Med.Chem., 51, 2008
|
|
4BZO
 
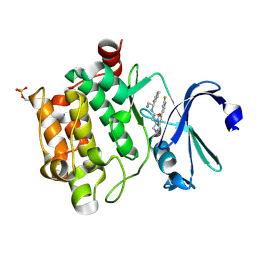 | | Crystal structure of PIM1 in complex with a Pyrrolo-Pyrazinone inhibitor | | 分子名称: | N-[(1S)-2-AMINO-1-PHENYLETHYL]-2-[(4S)-7-(2-FLUORO-4-PYRIDINYL)-1-OXO-1,2,3,4-TETRAHYDROPYRROLO[1,2-A]PYRAZIN-4-YL]ACETAMIDE, SERINE/THREONINE-PROTEIN KINASE PIM-1 | | 著者 | Casale, E, Casuscelli, F, Ardini, E, Avanzi, N, Cervi, G, D'Anello, M, Donati, D, Faiardi, D, Ferguson, R.D, Fogliatto, G, Galvani, A, Marsiglio, A, Mirizzi, D.G, Montemartini, M, Orrenius, C, Papeo, G, Piutti, C, Salom, B, Felder, E.R. | | 登録日 | 2013-07-29 | | 公開日 | 2013-10-30 | | 最終更新日 | 2013-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Discovery and Optimization of Pyrrolo[1,2-A]Pyrazinones Leads to Novel and Selective Inhibitors of Pim Kinases.
Bioorg.Med.Chem., 21, 2013
|
|
4BZN
 
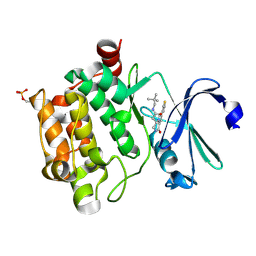 | | Crystal structure of PIM1 in complex with a Pyrrolo(1,2-a)Pyrazinone inhibitor | | 分子名称: | N-(2,2-dimethylpropyl)-2-[1-oxo-7-(thiophen-3-yl)-1,2,3,4-tetrahydropyrrolo[1,2-a]pyrazin-4-yl]acetamide, SERINE/THREONINE-PROTEIN KINASE PIM-1 | | 著者 | Casale, E, Casuscelli, F, Ardini, E, Avanzi, N, Cervi, G, D'Anello, M, Donati, D, Faiardi, D, Ferguson, R.D, Fogliatto, G, Galvani, A, Marsiglio, A, Mirizzi, D.G, Montemartini, M, Orrenius, C, Papeo, G, Piutti, C, Salom, B, Felder, E.R. | | 登録日 | 2013-07-29 | | 公開日 | 2013-10-30 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Discovery and Optimization of Pyrrolo[1,2-A]Pyrazinones Leads to Novel and Selective Inhibitors of Pim Kinases.
Bioorg.Med.Chem., 21, 2013
|
|
4Q0P
 
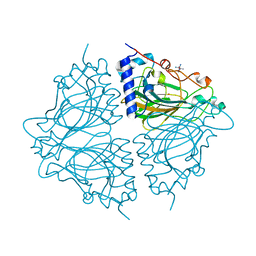 | | Crystal structure of Acinetobacter sp. DL28 L-ribose isomerase in complex with L-ribose | | 分子名称: | COBALT (II) ION, COBALT HEXAMMINE(III), L-Ribose isomerase, ... | | 著者 | Yoshida, H, Yoshihara, A, Teraoka, M, Izumori, K, Kamitori, S. | | 登録日 | 2014-04-02 | | 公開日 | 2014-05-28 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | X-ray structure of a novel L-ribose isomerase acting on a non-natural sugar L-ribose as its ideal substrate.
Febs J., 281, 2014
|
|
4Q0S
 
 | | Crystal structure of Acinetobacter sp. DL28 L-ribose isomerase in complex with ribitol | | 分子名称: | COBALT (II) ION, COBALT HEXAMMINE(III), D-ribitol, ... | | 著者 | Yoshida, H, Yoshihara, A, Teraoka, M, Izumori, K, Kamitori, S. | | 登録日 | 2014-04-02 | | 公開日 | 2014-05-28 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | X-ray structure of a novel L-ribose isomerase acting on a non-natural sugar L-ribose as its ideal substrate.
Febs J., 281, 2014
|
|
4D61
 
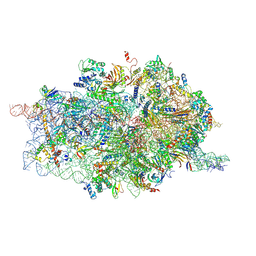 | | Cryo-EM structures of ribosomal 80S complexes with termination factors and cricket paralysis virus IRES reveal the IRES in the translocated state | | 分子名称: | 18S RRNA, 40S RIBOSOMAL PROTEIN S10, 40S RIBOSOMAL PROTEIN S11, ... | | 著者 | Muhs, M, Hilal, T, Mielke, T, Skabkin, M.A, Sanbonmatsu, K.Y, Pestova, T.V, Spahn, C.M.T. | | 登録日 | 2014-11-07 | | 公開日 | 2015-03-04 | | 最終更新日 | 2017-08-30 | | 実験手法 | ELECTRON MICROSCOPY (9 Å) | | 主引用文献 | Cryo-Em of Ribosomal 80S Complexes with Termination Factors Reveals the Translocated Cricket Paralysis Virus Ires.
Mol.Cell, 57, 2015
|
|
2ERK
 
 | |
