8DF0
 
 | |
6ECI
 
 | | Structure of the FAD binding protein MSMEG_5243 from Mycobacterium smegmatis | | 分子名称: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Pyridoxamine 5'-phosphate oxidase-related, ... | | 著者 | Ahmed, F.H, Antoney, J, Carr, P.D, Jackson, C.J. | | 登録日 | 2018-08-07 | | 公開日 | 2018-12-26 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | FAD-sequestering proteins protect mycobacteria against hypoxic and oxidative stress.
J. Biol. Chem., 294, 2019
|
|
8B0F
 
 | | CryoEM structure of C5b8-CD59 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Bubeck, D, Couves, E.C, Gardner, S. | | 登録日 | 2022-09-07 | | 公開日 | 2023-02-22 | | 最終更新日 | 2023-03-01 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis for membrane attack complex inhibition by CD59.
Nat Commun, 14, 2023
|
|
8B0G
 
 | | 2C9, C5b9-CD59 structure | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD59 glycoprotein, Complement C5, ... | | 著者 | Couves, E.C, Gardner, S, Bubeck, D. | | 登録日 | 2022-09-07 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural basis for membrane attack complex inhibition by CD59.
Nat Commun, 14, 2023
|
|
7ORW
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00265 | | 分子名称: | 1H-benzimidazol-4-amine, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | 登録日 | 2021-06-06 | | 公開日 | 2021-10-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
7ORU
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00221 | | 分子名称: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | 登録日 | 2021-06-06 | | 公開日 | 2021-10-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
2J3D
 
 | |
7ORV
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00239 | | 分子名称: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | 登録日 | 2021-06-06 | | 公開日 | 2021-10-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
7ORR
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00022 | | 分子名称: | 4-PHENYL-1H-IMIDAZOLE, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | 登録日 | 2021-06-06 | | 公開日 | 2021-10-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
5MQF
 
 | | Cryo-EM structure of a human spliceosome activated for step 2 of splicing (C* complex) | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, ATP-dependent RNA helicase DHX8, Cell division cycle 5-like protein, ... | | 著者 | Bertram, K, Hartmuth, K, Kastner, B. | | 登録日 | 2016-12-20 | | 公開日 | 2017-03-22 | | 最終更新日 | 2018-11-21 | | 実験手法 | ELECTRON MICROSCOPY (5.9 Å) | | 主引用文献 | Cryo-EM structure of a human spliceosome activated for step 2 of splicing.
Nature, 542, 2017
|
|
7OJN
 
 | | Lassa virus L protein in an elongation conformation [ELONGATION] | | 分子名称: | 3' RNA, 5' RNA, 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, ... | | 著者 | Kouba, T, Vogel, D, Thorkelsson, S, Quemin, E, Williams, H.M, Milewski, M, Busch, C, Gunther, S, Grunewald, K, Rosenthal, M, Cusack, S. | | 登録日 | 2021-05-16 | | 公開日 | 2021-12-01 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.92 Å) | | 主引用文献 | Conformational changes in Lassa virus L protein associated with promoter binding and RNA synthesis activity.
Nat Commun, 12, 2021
|
|
1LEM
 
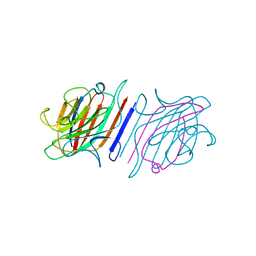 | |
3QZV
 
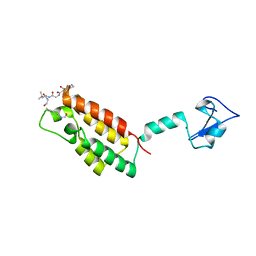 | | Crystal Structure of BPTF PHD-linker-bromo in complex with histone H4K12ac peptide | | 分子名称: | Histone H4, Nucleosome-remodeling factor subunit BPTF, ZINC ION | | 著者 | Li, H, Ruthenburg, A.J, Patel, D.J. | | 登録日 | 2011-03-07 | | 公開日 | 2011-06-01 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.999 Å) | | 主引用文献 | Recognition of a Mononucleosomal Histone Modification Pattern by BPTF via Multivalent Interactions.
Cell(Cambridge,Mass.), 145, 2011
|
|
6F6O
 
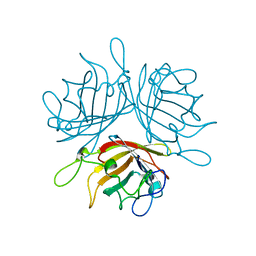 | |
8DWH
 
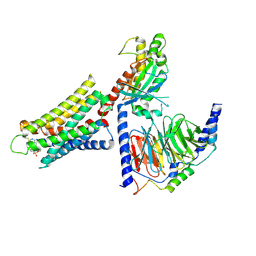 | | CryoEM structure of Gq-coupled MRGPRX1 with ligand Compound-16 | | 分子名称: | Gs-mini-Gq chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Liu, Y, Cao, C, Fay, J.F, Roth, B.L. | | 登録日 | 2022-08-01 | | 公開日 | 2022-11-02 | | 最終更新日 | 2023-04-12 | | 実験手法 | ELECTRON MICROSCOPY (3.25 Å) | | 主引用文献 | Ligand recognition and allosteric modulation of the human MRGPRX1 receptor.
Nat.Chem.Biol., 19, 2023
|
|
6F1T
 
 | | Cryo-EM structure of two dynein tail domains bound to dynactin and BICDR1 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ARP1 actin related protein 1 homolog A, ... | | 著者 | Urnavicius, L, Lau, C.K, Elshenawy, M.M, Morales-Rios, E, Motz, C, Yildiz, A, Carter, A.P. | | 登録日 | 2017-11-23 | | 公開日 | 2018-01-17 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Cryo-EM shows how dynactin recruits two dyneins for faster movement.
Nature, 554, 2018
|
|
8DWC
 
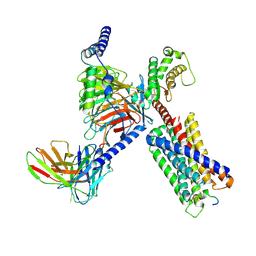 | | CryoEM structure of Gq-coupled MRGPRX1 with peptide agonist BAM8-22 | | 分子名称: | Gs-mini-Gq chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Liu, Y, Cao, C, Fay, J.F, Roth, B.L. | | 登録日 | 2022-08-01 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (2.87 Å) | | 主引用文献 | Ligand recognition and allosteric modulation of the human MRGPRX1 receptor.
Nat.Chem.Biol., 19, 2023
|
|
6BN8
 
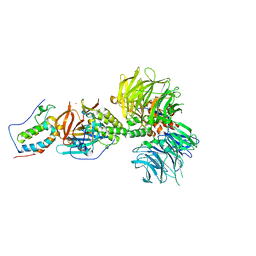 | | Crystal structure of DDB1-CRBN-BRD4(BD1) complex bound to dBET55 PROTAC. | | 分子名称: | Bromodomain-containing protein 4, DNA damage-binding protein 1,DNA damage-binding protein 1, Protein cereblon, ... | | 著者 | Nowak, R.P, DeAngelo, S.L, Buckley, D, Bradner, J.E, Fischer, E.S. | | 登録日 | 2017-11-16 | | 公開日 | 2018-06-06 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.990035 Å) | | 主引用文献 | Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nat. Chem. Biol., 14, 2018
|
|
8DWG
 
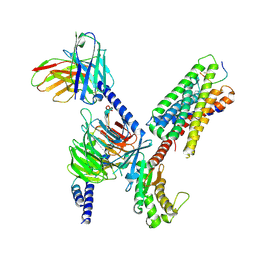 | | CryoEM structure of Gq-coupled MRGPRX1 with peptide ligand BAM8-22 and positive allosteric modulator ML382 | | 分子名称: | 2-[(cyclopropanesulfonyl)amino]-N-(2-ethoxyphenyl)benzamide, Gs-mini-Gq chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Liu, Y, Cao, C, Fay, J.F, Roth, B.L. | | 登録日 | 2022-08-01 | | 公開日 | 2022-11-02 | | 最終更新日 | 2023-04-12 | | 実験手法 | ELECTRON MICROSCOPY (2.71 Å) | | 主引用文献 | Ligand recognition and allosteric modulation of the human MRGPRX1 receptor.
Nat.Chem.Biol., 19, 2023
|
|
6FF7
 
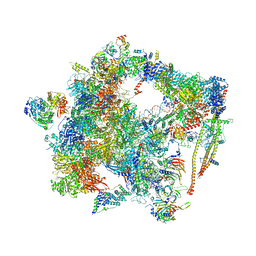 | | human Bact spliceosome core structure | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-DIPHOSPHATE, BUD13 homolog, ... | | 著者 | Haselbach, D, Komarov, I, Agafonov, D, Hartmuth, K, Graf, B, Kastner, B, Luehrmann, R, Stark, H. | | 登録日 | 2018-01-03 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Structure and Conformational Dynamics of the Human Spliceosomal BactComplex.
Cell, 172, 2018
|
|
6F3A
 
 | | Cryo-EM structure of a single dynein tail domain bound to dynactin and BICD2N | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ARP1 actin related protein 1 homolog A, ... | | 著者 | Urnavicius, L, Lau, C.K, Elshenawy, M.M, Morales-Rios, E, Motz, C, Yildiz, A, Carter, A.P. | | 登録日 | 2017-11-28 | | 公開日 | 2018-01-17 | | 最終更新日 | 2019-12-11 | | 実験手法 | ELECTRON MICROSCOPY (8.2 Å) | | 主引用文献 | Cryo-EM shows how dynactin recruits two dyneins for faster movement.
Nature, 554, 2018
|
|
3RUY
 
 | | Crystal Structure of the Ornithine-oxo acid transaminase RocD from Bacillus anthracis | | 分子名称: | Ornithine aminotransferase | | 著者 | Anderson, S.M, Wawrzak, Z, Brunzelle, J.S, Onopriyenko, O, Peterson, S.N, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-05-05 | | 公開日 | 2011-05-18 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Crystal Structure of the Ornithine-oxo acid transaminase RocD from Bacillus anthracis
To be Published
|
|
6EWG
 
 | |
6F29
 
 | | Crystal structure of the kainate receptor GluK3 ligand binding domain in complex with (S)-1-[2-Amino-2-carboxyethyl]-5,7-dihydrothieno[3,4-d]pyrimidin-2,4(1H,3H)-dione at resolution 2.6A | | 分子名称: | (2~{S})-2-azanyl-3-[2,4-bis(oxidanylidene)-5,7-dihydrothieno[3,4-d]pyrimidin-1-yl]propanoic acid, CHLORIDE ION, Glutamate receptor ionotropic, ... | | 著者 | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | 登録日 | 2017-11-23 | | 公開日 | 2018-02-28 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | ( S)-2-Amino-3-(5-methyl-3-hydroxyisoxazol-4-yl)propanoic Acid (AMPA) and Kainate Receptor Ligands: Further Exploration of Bioisosteric Replacements and Structural and Biological Investigation.
J. Med. Chem., 61, 2018
|
|
1XX5
 
 | | Crystal Structure of Natrin from Naja atra snake venom | | 分子名称: | ETHANOL, Natrin 1 | | 著者 | Wang, J, Shen, B, Lou, X.H, Guo, M, Teng, M.K, Niu, L.W. | | 登録日 | 2004-11-04 | | 公開日 | 2005-06-14 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Blocking effect and crystal structure of natrin toxin, a cysteine-rich secretory protein from Naja atra venom that targets the BKCa channel
Biochemistry, 44, 2005
|
|
