8A1S
 
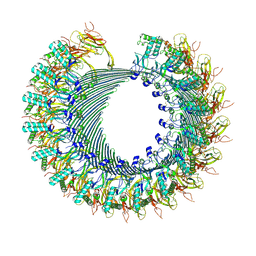 | | Structure of murine perforin-2 (Mpeg1) pore in twisted form | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Macrophage-expressed gene 1 protein | | 著者 | Yu, X, Ni, T, Zhang, P, Gilbert, R. | | 登録日 | 2022-06-02 | | 公開日 | 2022-07-20 | | 最終更新日 | 2023-10-18 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Cryo-EM structures of perforin-2 in isolation and assembled on a membrane suggest a mechanism for pore formation.
Embo J., 41, 2022
|
|
8A1D
 
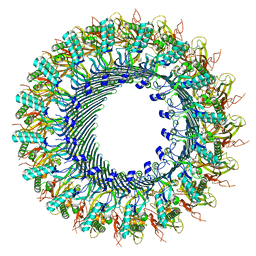 | | Structure of murine perforin-2 (Mpeg1) pore in ring form | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE, Macrophage-expressed gene 1 protein | | 著者 | Yu, X, Ni, T, Zhang, P, Gilbert, R. | | 登録日 | 2022-06-01 | | 公開日 | 2022-07-20 | | 最終更新日 | 2022-12-14 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Cryo-EM structures of perforin-2 in isolation and assembled on a membrane suggest a mechanism for pore formation.
Embo J., 41, 2022
|
|
6SB5
 
 | |
6OAR
 
 | |
6OAT
 
 | |
7ELK
 
 | |
5B61
 
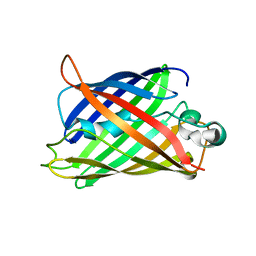 | | Extra-superfolder GFP | | 分子名称: | Green fluorescent protein | | 著者 | Park, H.H, Jang, T.-H, Choi, J.Y. | | 登録日 | 2016-05-24 | | 公開日 | 2017-06-14 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.115 Å) | | 主引用文献 | The mechanism of folding robustness revealed by the crystal structure of extra-superfolder GFP.
FEBS Lett., 591, 2017
|
|
5A8L
 
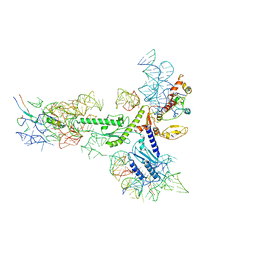 | | Human eRF1 and the hCMV nascent peptide in the translation termination complex | | 分子名称: | 28S RIBOSOMAL RNA, 60S RIBOSOMAL PROTEIN L12, 60S RIBOSOMAL PROTEIN L17, ... | | 著者 | Matheisl, S, Berninghausen, O, Becker, T, Beckmann, R. | | 登録日 | 2015-07-16 | | 公開日 | 2015-12-02 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structure of a Human Translation Termination Complex.
Nucleic Acids Res., 43, 2015
|
|
8U57
 
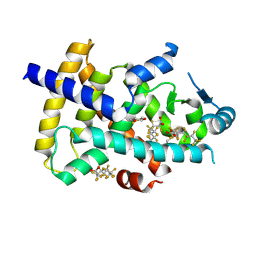 | | PPARg LBD in complex with perfluorooctanoic acid (PFOA) | | 分子名称: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, Peroxisome proliferator-activated receptor gamma, pentadecafluorooctanoic acid | | 著者 | Pederick, J.L, Frkic, R.L, McDougal, D.P, Bruning, J.B. | | 登録日 | 2023-09-12 | | 公開日 | 2024-07-24 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A structural basis for the activation of peroxisome proliferator-activated receptor gamma (PPAR gamma ) by perfluorooctanoic acid (PFOA).
Chemosphere, 354, 2024
|
|
5HU6
 
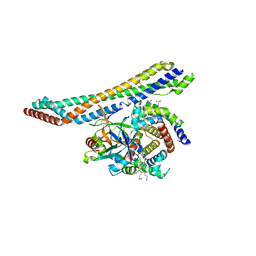 | |
5JDO
 
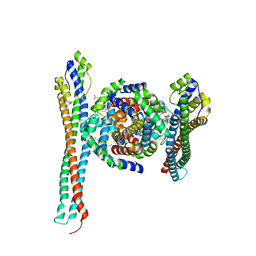 | |
7NDP
 
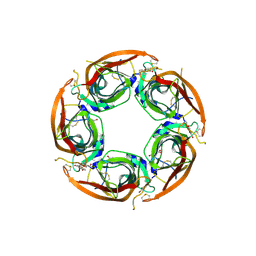 | | X-ray structure of acetylcholine-binding protein (AChBP) in complex with FL001856. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-bromanylspiro[3~{H}-chromene-2,4'-piperidine]-4-one, ... | | 著者 | Cederfelt, D, Boronat, P, Dobritzsch, D, Hennig, S, Fitzgerald, E.A, de Esch, I.J.P, Danielson, U.H. | | 登録日 | 2021-02-02 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Discovery of fragments inducing conformational effects in dynamic proteins using a second-harmonic generation biosensor.
Rsc Adv, 11, 2021
|
|
7AAI
 
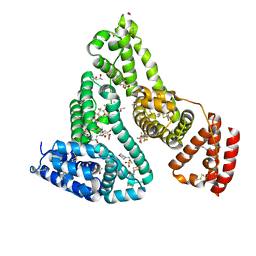 | | Crystal structure of Human serum albumin in complex with perfluorooctanoic acid (PFOA) at 2.10 Angstrom Resolution | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ... | | 著者 | Maso, L, Liberi, S, Trande, M, Angelini, A, Cendron, L. | | 登録日 | 2020-09-04 | | 公開日 | 2021-02-24 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Unveiling the binding mode of perfluorooctanoic acid to human serum albumin.
Protein Sci., 30, 2021
|
|
3D30
 
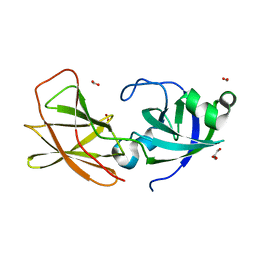 | | Structure of an expansin like protein from Bacillus Subtilis at 1.9A resolution | | 分子名称: | Expansin like protein, FORMIC ACID, GLYCEROL | | 著者 | Kerff, F, Petrella, S, Herman, R, Sauvage, E, Joris, B, Charlier, P. | | 登録日 | 2008-05-09 | | 公開日 | 2008-10-14 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure and activity of Bacillus subtilis YoaJ (EXLX1), a bacterial expansin that promotes root colonization.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3D2Z
 
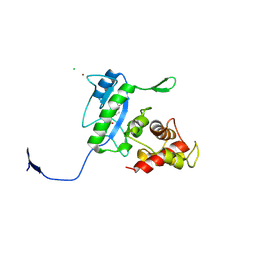 | | Complex of the N-acetylmuramyl-L-alanine amidase AmiD from E.coli with the product L-Ala-D-gamma-Glu-L-Lys | | 分子名称: | CHLORIDE ION, L-Ala-D-gamma-Glu-L-Lys peptide, N-acetylmuramoyl-L-alanine amidase amiD, ... | | 著者 | Kerff, F, Petrella, S, Herman, R, Sauvage, E, Mercier, F, Luxen, A, Frere, J.M, Joris, B, Charlier, P. | | 登録日 | 2008-05-09 | | 公開日 | 2009-06-16 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Specific Structural Features of the N-Acetylmuramoyl-l-Alanine Amidase AmiD from Escherichia coli and Mechanistic Implications for Enzymes of This Family.
J.Mol.Biol., 397, 2010
|
|
3D2Y
 
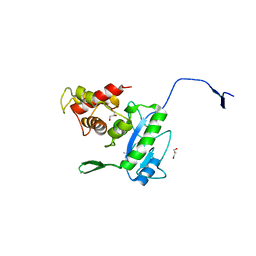 | | Complex of the N-acetylmuramyl-L-alanine amidase AmiD from E.coli with the substrate anhydro-N-acetylmuramic acid-L-Ala-D-gamma-Glu-L-Lys | | 分子名称: | Anhydro-N-acetylmuramic acid-L-Ala-D-gamma-Glu-L-Lys, GLYCEROL, N-acetylmuramoyl-L-alanine amidase amiD | | 著者 | Kerff, F, Petrella, S, Herman, R, Sauvage, E, Mercier, F, Luxen, A, Frere, J.M, Joris, B, Charlier, P. | | 登録日 | 2008-05-09 | | 公開日 | 2009-06-16 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Specific Structural Features of the N-Acetylmuramoyl-l-Alanine Amidase AmiD from Escherichia coli and Mechanistic Implications for Enzymes of This Family.
J.Mol.Biol., 397, 2010
|
|
6NUS
 
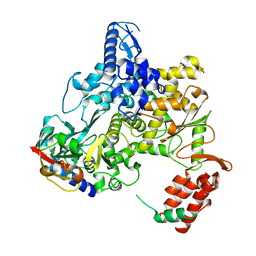 | |
6NUR
 
 | |
6NUT
 
 | |
6GT9
 
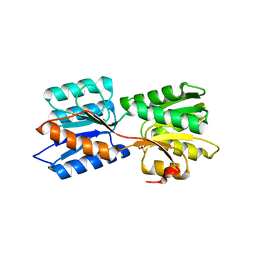 | | Crystal structure of GanP, a glucose-galactose binding protein from Geobacillus stearothermophilus, in complex with galactose | | 分子名称: | Putative sugar binding protein, SULFATE ION, beta-D-galactopyranose | | 著者 | Sherf, D, Lansky, S, Zehavi, A, Shoham, Y, Shoham, G. | | 登録日 | 2018-06-16 | | 公開日 | 2019-06-26 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.894 Å) | | 主引用文献 | The crystal structure of GanP, a glucose-galactose binding protein from Geobacillus stearothermophilus, in complex with galactose
To Be Published
|
|
5I08
 
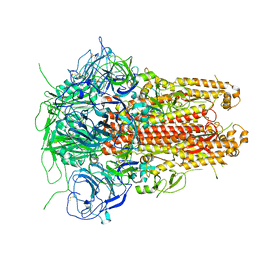 | | Prefusion structure of a human coronavirus spike protein | | 分子名称: | Spike glycoprotein,Foldon chimera | | 著者 | Kirchdoerfer, R.N, Cottrell, C.A, Wang, N, Pallesen, J, Yassine, H.M, Turner, H.L, Corbett, K.S, Graham, B.S, McLellan, J.S, Ward, A.B. | | 登録日 | 2016-02-03 | | 公開日 | 2016-03-02 | | 最終更新日 | 2020-04-22 | | 実験手法 | ELECTRON MICROSCOPY (4.04 Å) | | 主引用文献 | Pre-fusion structure of a human coronavirus spike protein.
Nature, 531, 2016
|
|
6GQ0
 
 | | Crystal structure of GanP, a glucose-galactose binding protein from Geobacillus stearothermophilus | | 分子名称: | Putative sugar binding protein | | 著者 | Sherf, D, Lansky, S, Zehavi, A, Shoham, Y, Shoham, G. | | 登録日 | 2018-06-07 | | 公開日 | 2019-06-19 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | The crystal structure of GanP, a glucose-galactose binding protein from Gebacillus Stearothermophilus
To Be Published
|
|
6GUQ
 
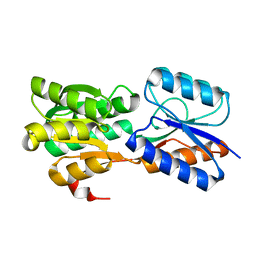 | | Crystal structure of GanP, a glucose-galactose binding protein from Geobacillus stearothermophilus, in complex with glucose | | 分子名称: | Putative sugar binding protein, beta-D-glucopyranose | | 著者 | Sherf, D, Lansky, S, Zehavi, A, Shoham, Y, Shoham, G. | | 登録日 | 2018-06-19 | | 公開日 | 2019-07-03 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.385 Å) | | 主引用文献 | The crystal structure of GanP, a glucose-galactose binding protein from Geobacillus stearothermophilus, in complex with glucose
To Be Published
|
|
2DIX
 
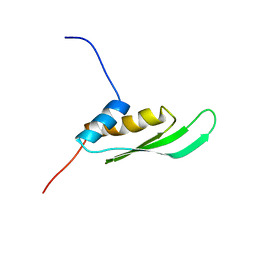 | | Solution structure of the DSRM domain of Protein activator of the interferon-induced protein kinase | | 分子名称: | Interferon-inducible double stranded RNA-dependent protein kinase activator A | | 著者 | Dang, W, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2006-03-30 | | 公開日 | 2006-09-30 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the DSRM domain of Protein activator of the interferon-induced protein kinase
To be published
|
|
4N6X
 
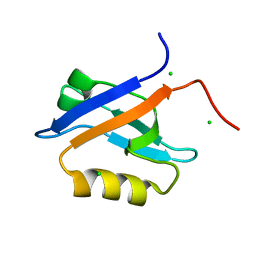 | | Crystal Structure of the Chemokine Receptor CXCR2 in Complex with the First PDZ Domain of NHERF1 | | 分子名称: | CHLORIDE ION, Na(+)/H(+) exchange regulatory cofactor NHE-RF1/Chemokine Receptor CXCR2 fusion protein | | 著者 | Lu, G, Wu, Y, Jiang, Y, Brunzelle, J, Sirinupong, N, Li, C, Yang, Z. | | 登録日 | 2013-10-14 | | 公開日 | 2014-01-15 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.051 Å) | | 主引用文献 | New Conformational State of NHERF1-CXCR2 Signaling Complex Captured by Crystal Lattice Trapping.
Plos One, 8, 2013
|
|
