5NHL
 
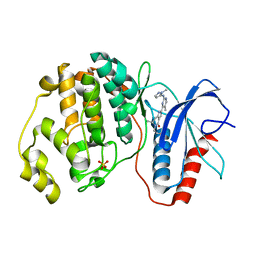 | | Human Erk2 with an Erk1/2 inhibitor | | Descriptor: | (6~{R})-5-(2-methoxyethyl)-6-methyl-2-[5-methyl-2-[(2-methylpyrazol-3-yl)amino]pyrimidin-4-yl]-6,7-dihydro-1~{H}-pyrrolo[3,2-c]pyridin-4-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Debreczeni, J.E, Ward, R.A, Bethel, P, Cook, C, Davies, E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, Hudson, J, James, M, Jones, C.D, Jones, C.R, Lamont, S, Lewis, R, Lindsay, N, Roberts, K, Simpson, I, StGallay, S, Swallow, S, Tonge, M. | | Deposit date: | 2017-03-21 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective Inhibitors of ERK1/2 from a Modestly Active and Promiscuous Chemical Start Point.
J. Med. Chem., 60, 2017
|
|
4AQ2
 
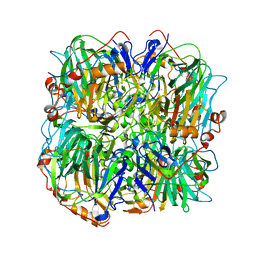 | | resting state of homogentisate 1,2-dioxygenase | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE (III) ION, HOMOGENTISATE 1,2-DIOXYGENASE | | Authors: | Jeoung, J.-H, Lin, T.-Y, Bommer, M, Dobbek, H. | | Deposit date: | 2012-04-12 | | Release date: | 2013-04-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Visualizing the Substrate-, Superoxo-, Alkylperoxo-, and Product-Bound States at the Nonheme Fe(II) Site of Homogentisate Dioxygenase.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7SEP
 
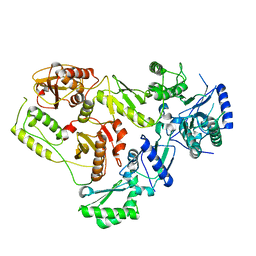 | |
6V7Y
 
 | | Human CD1d presenting alpha-Galactosylceramide in complex with VHH nanobody 1D5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d, ... | | Authors: | Shahine, A, Rossjohn, J. | | Deposit date: | 2019-12-10 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A single-domain bispecific antibody targeting CD1d and the NKT T-cell receptor induces a potent antitumor response.
Nat Cancer, 2020
|
|
5B7A
 
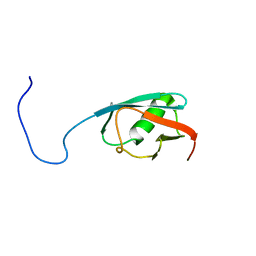 | |
1TZW
 
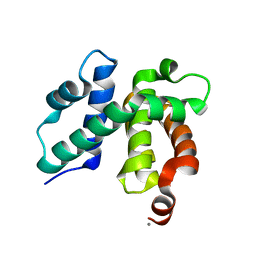 | | T. maritima NusB, P3121, Form 2 | | Descriptor: | CALCIUM ION, N utilization substance protein B homolog | | Authors: | Bonin, I, Robelek, R, Benecke, H, Urlaub, H, Bacher, A, Richter, G, Wahl, M.C. | | Deposit date: | 2004-07-12 | | Release date: | 2004-08-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of the antitermination factor NusB from Thermotoga maritima and implications for RNA binding
Biochem.J., 383, 2004
|
|
4AQ6
 
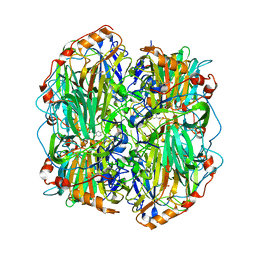 | | substrate bound homogentisate 1,2-dioxygenase | | Descriptor: | 2-(3,6-DIHYDROXYPHENYL)ACETIC ACID, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE (III) ION, ... | | Authors: | Jeoung, J.-H, Lin, T.-Y, Bommer, M, Dobbek, H. | | Deposit date: | 2012-04-12 | | Release date: | 2013-04-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Visualizing the Substrate-, Superoxo-, Alkylperoxo- and Product-Bound States at the Non-Heme Fe(II) Site of Homogentisate Dioxygenase
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4ALV
 
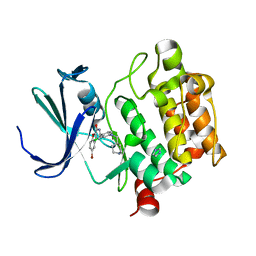 | | Benzofuropyrimidinone Inhibitors of Pim-1 | | Descriptor: | 8-bromo-2-{2-chloro-4-[(piperidin-4-ylmethyl)amino]phenyl}[1]benzofuro[3,2-d]pyrimidin-4(3H)-one, IMIDAZOLE, SERINE/THREONINE-PROTEIN KINASE PIM-1 | | Authors: | Stout, T.J, Adams, L. | | Deposit date: | 2012-03-05 | | Release date: | 2013-01-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | The Design, Synthesis, and Biological Evaluation of Pim Kinase Inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3ZDS
 
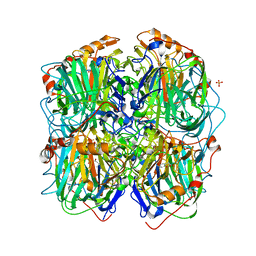 | | Structure of homogentisate 1,2-dioxygenase in complex with reaction intermediates of homogentisate with oxygen. | | Descriptor: | 2-(3,6-DIHYDROXYPHENYL)ACETIC ACID, 2-(6-oxidanyl-3-oxidanylidene-cyclohexa-1,4-dien-1-yl)ethanoic acid, 2-[(6R)-6-(dioxidanyl)-6-oxidanyl-3-oxidanylidene-cyclohexa-1,4-dien-1-yl]ethanoic acid, ... | | Authors: | Jeoung, J.-H, Bommer, M, Lin, T.-Y, Dobbek, H. | | Deposit date: | 2012-11-30 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Visualizing the Substrate-, Superoxo-, Alkylperoxo- and Product-Bound States at the Non-Heme Fe(II) Site of Homogentisate Dioxygenase
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2Q1Z
 
 | |
1MY7
 
 | | NF-kappaB p65 subunit dimerization domain homodimer N202R mutation | | Descriptor: | NF-kappaB p65 (RelA) subunit | | Authors: | Huxford, T, Mishler, D, Phelps, C.B, Huang, D.-B, Sengchanthalangsy, L.L, Reeves, R, Hughes, C.A, Komives, E.A, Ghosh, G. | | Deposit date: | 2002-10-03 | | Release date: | 2002-12-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Solvent exposed non-contacting amino acids play a critical role in NF-kappaB/IkappaB alpha complex formation
J.Mol.Biol., 324, 2002
|
|
4IJM
 
 | | Crystal structure of circadian clock protein KaiC A422V mutant | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase KaiC, MAGNESIUM ION, ... | | Authors: | Egli, M, Pattanayek, R. | | Deposit date: | 2012-12-21 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.352 Å) | | Cite: | Loop-Loop Interactions Regulate KaiA-Stimulated KaiC Phosphorylation in the Cyanobacterial KaiABC Circadian Clock.
Biochemistry, 52, 2013
|
|
3QAO
 
 | | The crystal structure of the N-terminal domain of a MerR-like transcriptional regulator from Listeria monocytogenes EGD-e | | Descriptor: | GLYCEROL, MerR-like transcriptional regulator | | Authors: | Tan, K, Gu, M, Peterson, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-01-11 | | Release date: | 2011-01-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.874 Å) | | Cite: | The crystal structure of the N-terminal domain of a MerR-like transcriptional regulator from Listeria monocytogenes EGD-e
To be Published
|
|
5D8E
 
 | |
5UZ4
 
 | | The cryo-EM structure of YjeQ bound to the 30S subunit suggests a fidelity checkpoint function for this protein in ribosome assembly | | Descriptor: | 16S RIBOSOMAL RNA, 3'-O-(N-methylanthraniloyl)-beta:gamma-imidoguanosine-5'-triphosphate, 30S ribosomal protein S10, ... | | Authors: | Razi, A, Guarne, A, Ortega, J. | | Deposit date: | 2017-02-24 | | Release date: | 2017-04-19 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | The cryo-EM structure of YjeQ bound to the 30S subunit suggests a fidelity checkpoint function for this protein in ribosome assembly.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1A1D
 
 | | YEAST RNA POLYMERASE SUBUNIT RPB8, NMR, MINIMIZED AVERAGE STRUCTURE, ALPHA CARBONS ONLY | | Descriptor: | RNA POLYMERASE | | Authors: | Krapp, S, Kelly, G, Reischl, J, Weinzierl, R, Matthews, S. | | Deposit date: | 1997-12-10 | | Release date: | 1999-03-02 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Eukaryotic RNA polymerase subunit RPB8 is a new relative of the OB family.
Nat.Struct.Biol., 5, 1998
|
|
4ENX
 
 | | Crystal Structure of Pim-1 Kinase in complex with inhibitor (2E,5Z)-2-(2-chlorophenylimino)-5-(4-hydroxy-3-nitrobenzylidene)thiazolidin-4-one | | Descriptor: | (2Z,5Z)-2-[(2-chlorophenyl)imino]-5-(4-hydroxy-3-nitrobenzylidene)-1,3-thiazolidin-4-one, PHOSPHATE ION, Serine/threonine-protein kinase pim-1 | | Authors: | Parker, L.J, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-13 | | Release date: | 2012-08-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Flexibility of the P-loop of Pim-1 kinase: observation of a novel conformation induced by interaction with an inhibitor
Acta Crystallogr.,Sect.F, 68, 2012
|
|
8SOS
 
 | | Human CD1d presenting sphingomyelin C24:1 in complex with VHH nanobody 1D17 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d, Beta-2-microglobulin, ... | | Authors: | Shahine, A, Rossjohn, J. | | Deposit date: | 2023-04-29 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Enhanced CD1d phosphatidylserine presentation using a single-domain antibody promotes immunomodulatory CD1d-TIM-3 interactions.
J Immunother Cancer, 11, 2023
|
|
6Q7U
 
 | |
1E50
 
 | | AML1/CBFbeta complex | | Descriptor: | CORE-BINDING FACTOR ALPHA SUBUNIT, CORE-BINDING FACTOR CBF-BETA | | Authors: | Warren, A.J, Bravo, J, Williams, R.L, Rabbits, T.H. | | Deposit date: | 2000-07-13 | | Release date: | 2001-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for the Heterodimeric Interaction between the Acute Leukaemia-Associated Transcription Factors Aml1 and Cbfbeta
Embo J., 19, 2000
|
|
6CKO
 
 | | Crystal structure of an AF10 fragment | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, Protein AF-10, ... | | Authors: | Zhang, H, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-28 | | Release date: | 2018-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional analysis of the DOT1L-AF10 complex reveals mechanistic insights into MLL-AF10-associated leukemogenesis.
Genes Dev., 32, 2018
|
|
4OLT
 
 | | Chitosanase complex structure | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, Chitosanase, GLYCEROL | | Authors: | Liu, W.Z, Lyu, Q.Q, Han, B.Q. | | Deposit date: | 2014-01-25 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural insights into the substrate-binding mechanism for a novel chitosanase.
Biochem.J., 461, 2014
|
|
3R04
 
 | | The discovery of novel benzofuran-2-carboxylic acids as potent Pim-1 inhibitors | | Descriptor: | 5-{6-[(trans-4-aminocyclohexyl)amino]pyrazin-2-yl}-1-benzofuran-2-carboxylic acid, IMIDAZOLE, Proto-oncogene serine/threonine-protein kinase pim-1 | | Authors: | Xiang, Y, Hirth, B, Asmussen, G, Biemann, H.-P, Good, A, Fitzgerald, M, Gladysheva, T, Jancsics, K, Liu, J, Metz, M, Papoulis, A, Skerlj, R, Stepp, D.J, Wei, R.R. | | Deposit date: | 2011-03-07 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The discovery of novel benzofuran-2-carboxylic acids as potent Pim-1 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
6Q3G
 
 | | Structure of native bacteriophage P68 | | Descriptor: | Arstotzka protein, Head fiber protein, Inner core protein, ... | | Authors: | Dominik, H, Karel, S, Fuzik, T, Plevka, P. | | Deposit date: | 2018-12-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and genome ejection mechanism ofStaphylococcus aureusphage P68.
Sci Adv, 5, 2019
|
|
6D3A
 
 | | Structure of human ARH3 D314E bound to ADP-ribose and magnesium | | Descriptor: | MAGNESIUM ION, Poly(ADP-ribose) glycohydrolase ARH3, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Pourfarjam, Y, Ventura, J, Kurinov, I, Kim, I.K. | | Deposit date: | 2018-04-15 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.60001016 Å) | | Cite: | Structure of human ADP-ribosyl-acceptor hydrolase 3 bound to ADP-ribose reveals a conformational switch that enables specific substrate recognition.
J.Biol.Chem., 293, 2018
|
|
