1G0R
 
 | | THE STRUCTURAL BASIS OF THE CATALYTIC MECHANISM AND REGULATION OF GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE (RMLA). THYMIDINE/GLUCOSE-1-PHOSPHATE COMPLEX. | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, SULFATE ION, ... | | Authors: | Blankenfeldt, W, Asuncion, M, Lam, J.S, Naismith, J.H. | | Deposit date: | 2000-10-07 | | Release date: | 2000-12-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA).
EMBO J., 19, 2000
|
|
3GUS
 
 | | Crystal strcture of human Pi class glutathione S-transferase GSTP1-1 in complex with 6-(7-Nitro-2,1,3-benzoxadiazol-4-ylthio)hexanol (NBDHEX) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-[(7-nitro-2,1,3-benzoxadiazol-4-yl)sulfanyl]hexan-1-ol, GLUTATHIONE, ... | | Authors: | Federici, L, Lo Sterzo, C, Di Matteo, A, Scaloni, F, Federici, G, Caccuri, A.M. | | Deposit date: | 2009-03-30 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural basis for the binding of the anticancer compound 6-(7-nitro-2,1,3-benzoxadiazol-4-ylthio)hexanol to human glutathione s-transferases
Cancer Res., 69, 2009
|
|
4DQB
 
 | | Crystal Structure of wild-type HIV-1 Protease in Complex with DRV | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, 1,2-ETHANEDIOL, Aspartyl protease, ... | | Authors: | Schiffer, C.A, Mittal, S. | | Deposit date: | 2012-02-15 | | Release date: | 2012-03-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Hydrophobic core flexibility modulates enzyme activity in HIV-1 protease.
J.Am.Chem.Soc., 134, 2012
|
|
3NF7
 
 | | Structural basis for a new mechanism of inhibition of HIV integrase identified by fragment screening and structure based design | | Descriptor: | 1,2-ETHANEDIOL, 5-[(5-chloro-2-oxo-2,3-dihydro-1H-indol-1-yl)methyl]-1,3-benzodioxole-4-carboxylic acid, ACETIC ACID, ... | | Authors: | Peat, T.S, Newman, J, Deadman, J.J, Rhodes, D. | | Deposit date: | 2010-06-09 | | Release date: | 2011-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for a new mechanism of inhibition of HIV-1 integrase identified by fragment screening and structure-based design
ANTIVIR.CHEM.CHEMOTHER., 21, 2011
|
|
1TW5
 
 | | beta1,4-galactosyltransferase mutant M344H-Gal-T1 in complex with Chitobiose | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ramakrishnan, B, Boeggeman, E, Qasba, P.K. | | Deposit date: | 2004-06-30 | | Release date: | 2004-12-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Effect of the Met344His mutation on the conformational dynamics of bovine beta-1,4-galactosyltransferase: crystal structure of the Met344His mutant in complex with chitobiose
Biochemistry, 43, 2004
|
|
4KJX
 
 | | Crystal Structure of the complex of three phase partition treated lipase from Thermomyces lanuginosa with Lauric acid and P-nitrobenzaldehyde (PNB) at 2.1 resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-nitrobenzaldehyde, ... | | Authors: | Kumar, M, Mukherjee, J, Gupta, M.N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-05-04 | | Release date: | 2013-05-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the complex of three phase partition treated lipase from Thermomyces lanuginosa with Lauric acid and P-nitrobenzaldehyde (PNB) at 2.1 resolution
TO BE PUBLISHED
|
|
1ODY
 
 | | HIV-1 PROTEASE COMPLEXED WITH AN INHIBITOR LP-130 | | Descriptor: | 4-[2-(2-ACETYLAMINO-3-NAPHTALEN-1-YL-PROPIONYLAMINO)-4-METHYL-PENTANOYLAMINO]-3-HYDROXY-6-METHYL-HEPTANOIC ACID [1-(1-CARBAMOYL-2-NAPHTHALEN-1-YL-ETHYLCARBAMOYL)-PROPYL]-AMIDE, HIV-1 PROTEASE | | Authors: | Kervinen, J, Lubkowski, J, Zdanov, A, Wlodawer, A, Gustchina, A. | | Deposit date: | 1998-07-13 | | Release date: | 1999-02-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Toward a universal inhibitor of retroviral proteases: comparative analysis of the interactions of LP-130 complexed with proteases from HIV-1, FIV, and EIAV.
Protein Sci., 7, 1998
|
|
3OXC
 
 | | Wild Type HIV-1 Protease with Antiviral Drug Saquinavir | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, FORMIC ACID, Protease, ... | | Authors: | Kovalevsky, A.Y, Wang, Y.-F, Tie, Y, Weber, I.T. | | Deposit date: | 2010-09-21 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Atomic resolution crystal structures of HIV-1 protease and mutants V82A and I84V with saquinavir
Proteins, 67, 2007
|
|
3OXW
 
 | | Crystal Structure of HIV-1 I50V, A71V Protease in Complex with the Protease Inhibitor Darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Mittal, S, Bandaranayake, R.M, Schiffer, C.A. | | Deposit date: | 2010-09-22 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and thermodynamic basis of amprenavir/darunavir and atazanavir resistance in HIV-1 protease with mutations at residue 50.
J.Virol., 87, 2013
|
|
1K3Y
 
 | | Crystal Structure Analysis of human Glutathione S-transferase with S-hexyl glutatione and glycerol at 1.3 Angstrom | | Descriptor: | GLUTATHIONE S-TRANSFERASE A1, GLYCEROL, S-HEXYLGLUTATHIONE | | Authors: | Le Trong, I, Stenkamp, R.E, Ibarra, C, Atkins, W.M, Adman, E.T. | | Deposit date: | 2001-10-04 | | Release date: | 2002-10-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | 1.3-A resolution structure of human glutathione S-transferase with S-hexyl glutathione bound reveals possible extended ligandin binding site.
Proteins, 48, 2002
|
|
4AG9
 
 | | C. elegans glucosamine-6-phosphate N-acetyltransferase (GNA1): ternary complex with coenzyme A and GlcNAc | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, COENZYME A, ... | | Authors: | Dorfmueller, H.C, Fang, W, Rao, F.V, Blair, D.E, Attrill, H, Shepherd, S.M, van Aalten, D.M.F. | | Deposit date: | 2012-01-24 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and Biochemical Characterization of a Trapped Coenzyme a Adduct of Caenorhabditis Elegans Glucosamine-6-Phosphate N-Acetyltransferase 1.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3MXD
 
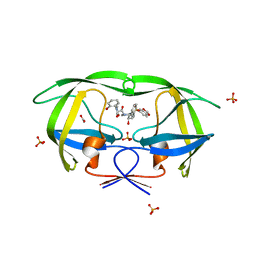 | | Crystal structure of HIV-1 protease inhibitor KC53 in complex with wild-type protease | | Descriptor: | (5S)-N-{(1S,2R)-3-[(1,3-benzodioxol-5-ylsulfonyl)(2-methylpropyl)amino]-1-benzyl-2-hydroxypropyl}-3-(2-hydroxyphenyl)-2 -oxo-1,3-oxazolidine-5-carboxamide, ACETATE ION, HIV-1 protease, ... | | Authors: | Nalam, M.N.L, Schiffer, C.A. | | Deposit date: | 2010-05-07 | | Release date: | 2010-11-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Based Design, Synthesis, and Structure-Activity Relationship Studies of HIV-1 Protease Inhibitors Incorporating Phenyloxazolidinones.
J.Med.Chem., 53, 2010
|
|
1WBM
 
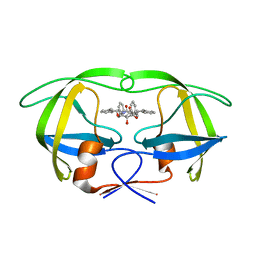 | | HIV-1 protease in complex with symmetric inhibitor, BEA450 | | Descriptor: | (2R,3R,4R,5R)-3,4-DIHYDROXY-N,N'-BIS[(1S,2R)-2-HYDROXY-2,3-DIHYDRO-1H-INDEN-1-YL]-2,5-BIS(2-PHENYLETHYL)HEXANEDIAMIDE, POL PROTEIN (FRAGMENT) | | Authors: | Lindberg, J, Unge, T. | | Deposit date: | 2004-11-02 | | Release date: | 2004-11-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | HIV-1 Protease in Complex with Symmetric Inhibitor, Bea450
To be Published
|
|
1SBG
 
 | |
4GE0
 
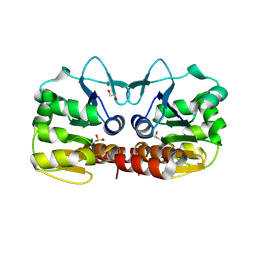 | | Schizosaccharomyces pombe DJ-1 T114P mutant | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Uncharacterized protein C22E12.03c | | Authors: | Madzelan, P, Labunska, T, Wilson, M.A. | | Deposit date: | 2012-08-01 | | Release date: | 2012-08-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Influence of peptide dipoles and hydrogen bonds on reactive cysteine pK(a) values in fission yeast DJ-1.
Febs J., 279, 2012
|
|
1MER
 
 | | HIV-1 MUTANT (I84V) PROTEASE COMPLEXED WITH DMP450 | | Descriptor: | HIV-1 PROTEASE, [4-R-(-4-ALPHA,5-ALPHA,6-BETA,7-BETA)]-HEXAHYDRO-5,6-BIS(HYDROXY)-1,3-BIS([(3-AMINO)PHENYL]METHYL)-4,7-BIS(PHENYLMETHYL)-2H-1,3-DIAZEPINONE | | Authors: | Ala, P, Chang, C.-H. | | Deposit date: | 1997-04-11 | | Release date: | 1998-04-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis of HIV-1 protease drug resistance: structural analysis of mutant proteases complexed with cyclic urea inhibitors.
Biochemistry, 36, 1997
|
|
3MXE
 
 | | Crystal structure of HIV-1 protease inhibitor, KC32 complexed with wild-type protease | | Descriptor: | (5S)-N-{(1S,2R)-3-[(1,3-benzothiazol-6-ylsulfonyl)(2-methylpropyl)amino]-1-benzyl-2-hydroxypropyl}-2-oxo-3-[2-(trifluoromethyl)phenyl]-1,3-oxazolidine-5-carboxamide, ACETATE ION, HIV-1 protease, ... | | Authors: | Nalam, M.N.L, Schiffer, C.A. | | Deposit date: | 2010-05-07 | | Release date: | 2010-11-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Based Design, Synthesis, and Structure-Activity Relationship Studies of HIV-1 Protease Inhibitors Incorporating Phenyloxazolidinones.
J.Med.Chem., 53, 2010
|
|
1MES
 
 | | HIV-1 MUTANT (I84V) PROTEASE COMPLEXED WITH DMP323 | | Descriptor: | HIV-1 PROTEASE, [4-R-(-4-ALPHA,5-ALPHA,6-BETA,7-BETA)]-HEXAHYDRO-5,6-BIS(HYDROXY)-[1,3-BIS([4-HYDROXYMETHYL-PHENYL]METHYL)-4,7-BIS(PHEN YLMETHYL)]-2H-1,3-DIAZEPINONE | | Authors: | Ala, P, Chang, C.-H. | | Deposit date: | 1997-04-11 | | Release date: | 1998-04-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis of HIV-1 protease drug resistance: structural analysis of mutant proteases complexed with cyclic urea inhibitors.
Biochemistry, 36, 1997
|
|
1HOS
 
 | |
1HQD
 
 | | PSEUDOMONAS CEPACIA LIPASE COMPLEXED WITH TRANSITION STATE ANALOGUE OF 1-PHENOXY-2-ACETOXY BUTANE | | Descriptor: | (RP,SP)-O-(2R)-(1-PHENOXYBUT-2-YL)-METHYLPHOSPHONIC ACID CHLORIDE, CALCIUM ION, LIPASE | | Authors: | Luic, M, Tomic, S, Lescic, I, Ljubovic, E, Sepac, D, Sunjic, V, Vitale, L, Saenger, W, Kojic-Prodic, B. | | Deposit date: | 2000-12-15 | | Release date: | 2001-08-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Complex of Burkholderia cepacia lipase with transition state analogue of 1-phenoxy-2-acetoxybutane: biocatalytic, structural and modelling study.
Eur.J.Biochem., 268, 2001
|
|
1HT6
 
 | | CRYSTAL STRUCTURE AT 1.5A RESOLUTION OF THE BARLEY ALPHA-AMYLASE ISOZYME 1 | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-AMYLASE ISOZYME 1, CALCIUM ION | | Authors: | Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2000-12-29 | | Release date: | 2003-07-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of barley alpha-amylase isozyme 1 reveals a novel role of domain C in substrate recognition and binding: a pair of sugar tongs
Structure, 11, 2003
|
|
4GB2
 
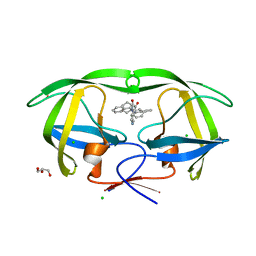 | | HIV-1 protease (mutant Q7K L33I L63I) in complex with a bicyclic pyrrolidine inhibitor | | Descriptor: | (4aS,7aS)-1,4-bis(diphenylmethyl)hexahydro-1H-pyrrolo[3,4-b]pyrazine-2,3-dione, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stieler, M, Heine, A, Klebe, G. | | Deposit date: | 2012-07-26 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.788 Å) | | Cite: | Cocrystallization of potent pyrrolidine based HIV-1 protease inhibitors
To be Published
|
|
1MTB
 
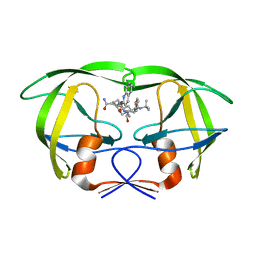 | | Viability of a drug-resistant HIV-1 protease mutant: structural insights for better antiviral therapy | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, PROTEASE RETROPEPSIN | | Authors: | Prabu-Jeyabalan, M, Nalivaika, E.A, King, N.M, Schiffer, C.A. | | Deposit date: | 2002-09-20 | | Release date: | 2003-01-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Viability of drug-resistant human immunodeficiency virus type 1 protease variant: structural insights for better antiviral therapy
J.Virol., 77, 2003
|
|
4GE3
 
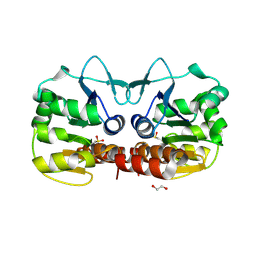 | | Schizosaccharomyces pombe DJ-1 T114V mutant | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Uncharacterized protein C22E12.03c | | Authors: | Madzelan, P, Labunska, T, Wilson, M.A. | | Deposit date: | 2012-08-01 | | Release date: | 2012-08-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Influence of peptide dipoles and hydrogen bonds on reactive cysteine pK(a) values in fission yeast DJ-1.
Febs J., 279, 2012
|
|
2G0A
 
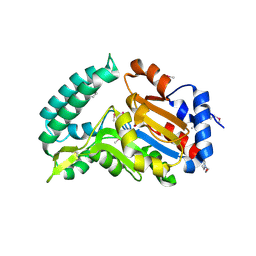 | | X-ray structure of mouse pyrimidine 5'-nucleotidase type 1 with lead(II) bound in active site | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Cytosolic 5'-nucleotidase III, LEAD (II) ION | | Authors: | Bitto, E, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-02-11 | | Release date: | 2006-04-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of pyrimidine 5'-nucleotidase type 1. Insight into mechanism of action and inhibition during lead poisoning.
J.Biol.Chem., 281, 2006
|
|
