2CY3
 
 | |
1DKA
 
 | | DIALKYLGLYCINE DECARBOXYLASE STRUCTURE: BIFUNCTIONAL ACTIVE SITE AND ALKALI METAL BINDING SITES | | Descriptor: | 2,2-DIALKYLGLYCINE DECARBOXYLASE (PYRUVATE), 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, POTASSIUM ION, ... | | Authors: | Toney, M.D, Hohenester, E, Jansonius, J.N. | | Deposit date: | 1993-06-18 | | Release date: | 1994-10-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Dialkylglycine decarboxylase structure: bifunctional active site and alkali metal sites.
Science, 261, 1993
|
|
2SAM
 
 | | STRUCTURE OF THE PROTEASE FROM SIMIAN IMMUNODEFICIENCY VIRUS: COMPLEX WITH AN IRREVERSIBLE NON-PEPTIDE INHIBITOR | | Descriptor: | 3-(4-NITRO-PHENOXY)-PROPAN-1-OL, SIV PROTEASE | | Authors: | Rose, R.B, Rose, J.R, Salto, R, Craik, C.S, Stroud, R.M. | | Deposit date: | 1994-07-08 | | Release date: | 1994-10-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the protease from simian immunodeficiency virus: complex with an irreversible nonpeptide inhibitor.
Biochemistry, 32, 1993
|
|
1HCD
 
 | | STRUCTURE OF HISACTOPHILIN IS SIMILAR TO INTERLEUKIN-1 BETA AND FIBROBLAST GROWTH FACTOR | | Descriptor: | HISACTOPHILIN | | Authors: | Habazettl, J, Gondol, D, Wiltscheck, R, Otlewski, J, Schleicher, M, Holak, T.A. | | Deposit date: | 1994-05-03 | | Release date: | 1994-10-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of hisactophilin is similar to interleukin-1 beta and fibroblast growth factor.
Nature, 359, 1992
|
|
1SBG
 
 | |
173D
 
 | | MULTIPLE BINDING MODES OF ANTICANCER DRUG ACTINOMYCIN D: X-RAY, MOLECULAR MODELING, AND SPECTROSCOPIC STUDIES OF D(GAAGCTTC)2-ACTINOMYCIN D COMPLEXES AND ITS HOST DNA | | Descriptor: | ACTINOMYCIN D, DNA (5'-D(*GP*AP*AP*GP*CP*TP*TP*C)-3') | | Authors: | Kamitori, S, Takusagawa, F. | | Deposit date: | 1994-04-18 | | Release date: | 1994-10-15 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Multiple Binding Modes of Anticancer Drug Actinomycin D: X-Ray, Molecular Modeling, and Spectroscopic Studies of D(Gaagcttc)2-Actinomycin D Complexes and its Host DNA
J.Am.Chem.Soc., 116, 1994
|
|
177D
 
 | |
1AZN
 
 | | CRYSTAL STRUCTURE OF THE AZURIN MUTANT PHE114ALA FROM PSEUDOMONAS AERUGINOSA AT 2.6 ANGSTROMS RESOLUTION | | Descriptor: | AZURIN, COPPER (II) ION | | Authors: | Tsai, L.-C, Sjolin, L, Langer, V, Pascher, T, Nar, H. | | Deposit date: | 1994-05-27 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the azurin mutant Phe114Ala from Pseudomonas aeruginosa at 2.6 A resolution.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1ERD
 
 | | THE NMR SOLUTION STRUCTURE OF THE PHEROMONE ER-2 FROM THE CILIATED PROTOZOAN EUPLOTES RAIKOVI | | Descriptor: | PHEROMONE ER-2 | | Authors: | Ottiger, M, Szyperski, T, Luginbuhl, P, Ortenzi, C, Luporini, P, Bradshaw, R.A, Wuthrich, K. | | Deposit date: | 1994-02-14 | | Release date: | 1994-10-15 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of the pheromone Er-2 from the ciliated protozoan Euplotes raikovi.
Protein Sci., 3, 1994
|
|
1BCX
 
 | |
2IRT
 
 | |
1AIB
 
 | |
1AIA
 
 | |
1ERC
 
 | | THE NMR SOLUTION STRUCTURE OF THE PHEROMONE ER-1 FROM THE CILIATED PROTOZOAN EUPLOTES RAIKOVI | | Descriptor: | PHEROMONE ER-1 | | Authors: | Mronga, S, Luginbuhl, P, Brown, L.R, Ortenzi, C, Luporini, P, Bradshaw, R.A, Wuthrich, K. | | Deposit date: | 1994-02-14 | | Release date: | 1994-10-15 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of the pheromone Er-1 from the ciliated protozoan Euplotes raikovi.
Protein Sci., 3, 1994
|
|
1ENJ
 
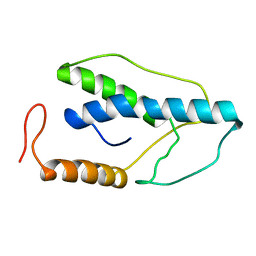 | | CRYSTAL STRUCTURE OF A PYRIMIDINE DIMER SPECIFIC EXCISION REPAIR ENZYME FROM BACTERIOPHAGE T4: REFINEMENT AT 1.45 ANGSTROMS AND X-RAY ANALYSIS OF THE THREE ACTIVE SITE MUTANTS | | Descriptor: | ENDONUCLEASE V | | Authors: | Vassylyev, D.G, Ariyoshi, M, Matsumoto, O, Katayanagi, K, Ohtsuka, E, Morikawa, K. | | Deposit date: | 1994-08-08 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a pyrimidine dimer-specific excision repair enzyme from bacteriophage T4: refinement at 1.45 A and X-ray analysis of the three active site mutants.
J.Mol.Biol., 249, 1995
|
|
1AIC
 
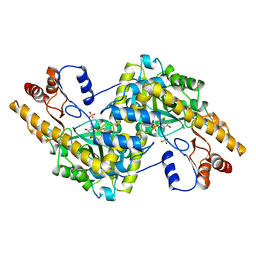 | |
1BGU
 
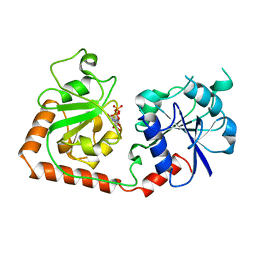 | | CRYSTAL STRUCTURE OF THE DNA MODIFYING ENZYME BETA-GLUCOSYLTRANSFERASE IN THE PRESENCE AND ABSENCE OF THE SUBSTRATE URIDINE DIPHOSPHOGLUCOSE | | Descriptor: | BETA-GLUCOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE | | Authors: | Vrielink, A, Rueger, W, Driessen, H.P.C, Freemont, P.S. | | Deposit date: | 1994-06-09 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the DNA modifying enzyme beta-glucosyltransferase in the presence and absence of the substrate uridine diphosphoglucose.
EMBO J., 13, 1994
|
|
1HRE
 
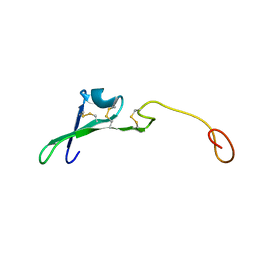 | | SOLUTION STRUCTURE OF THE EPIDERMAL GROWTH FACTOR-LIKE DOMAIN OF HEREGULIN-ALPHA, A LIGAND FOR P180ERB4 | | Descriptor: | HEREGULIN ALPHA | | Authors: | Nagata, K, Kohda, D, Hatanaka, H, Ichikawa, S, Inagaki, F. | | Deposit date: | 1994-07-21 | | Release date: | 1994-10-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the epidermal growth factor-like domain of heregulin-alpha, a ligand for p180erbB-4.
EMBO J., 13, 1994
|
|
1HVC
 
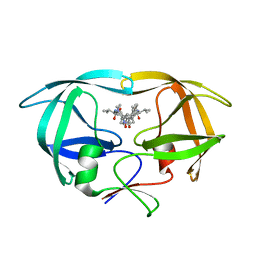 | | CRYSTAL STRUCTURE OF A TETHERED DIMER OF HIV-1 PROTEASE COMPLEXED WITH AN INHIBITOR | | Descriptor: | HIV-1 PROTEASE, N-{1-BENZYL-(2S,3S)-2,3-DIHYDROXY-4-[3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRYLAMINO]-5-PHENYL-PENTYL}-3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRAMIDE | | Authors: | Bhat, T.N, Baldwin, E.T, Erickson, J.W. | | Deposit date: | 1994-06-22 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a tethered dimer of HIV-1 proteinase complexed with an inhibitor.
Nat.Struct.Biol., 1, 1994
|
|
1NNT
 
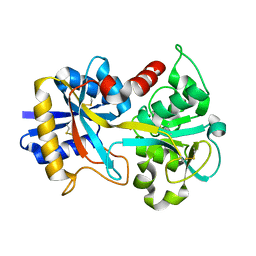 | |
1MNG
 
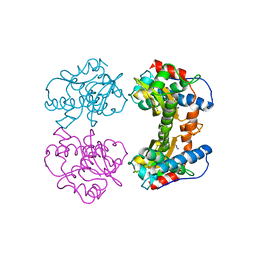 | | STRUCTURE-FUNCTION IN E. COLI IRON SUPEROXIDE DISMUTASE: COMPARISONS WITH THE MANGANESE ENZYME FROM T. THERMOPHILUS | | Descriptor: | AZIDE ION, MANGANESE (II) ION, MANGANESE SUPEROXIDE DISMUTASE | | Authors: | Lah, M.S, Dixon, M, Pattridge, K.A, Stallings, W.C, Fee, J.A, Ludwig, M.L. | | Deposit date: | 1994-07-13 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function in Escherichia coli iron superoxide dismutase: comparisons with the manganese enzyme from Thermus thermophilus.
Biochemistry, 34, 1995
|
|
1PAA
 
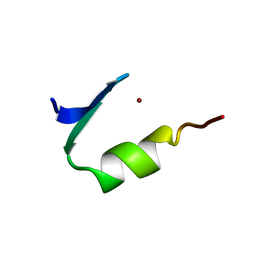 | | STRUCTURE OF A HISTIDINE-X4-HISTIDINE ZINC FINGER DOMAIN: INSIGHTS INTO ADR1-UAS1 PROTEIN-DNA RECOGNITION | | Descriptor: | YEAST TRANSCRIPTION FACTOR ADR1, ZINC ION | | Authors: | Bernstein, B.E, Hoffman, R.C, Horvath, S.J, Herriott, J.R, Klevit, R.E. | | Deposit date: | 1994-07-15 | | Release date: | 1994-10-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of a histidine-X4-histidine zinc finger domain: insights into ADR1-UAS1 protein-DNA recognition.
Biochemistry, 33, 1994
|
|
2DKB
 
 | | DIALKYLGLYCINE DECARBOXYLASE STRUCTURE: BIFUNCTIONAL ACTIVE SITE AND ALKALI METAL BINDING SITES | | Descriptor: | 2,2-DIALKYLGLYCINE DECARBOXYLASE (PYRUVATE), 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Toney, M.D, Hohenester, E, Jansonius, J.N. | | Deposit date: | 1994-07-12 | | Release date: | 1994-10-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dialkylglycine decarboxylase structure: bifunctional active site and alkali metal sites.
Science, 261, 1993
|
|
1TUS
 
 | |
3HVT
 
 | | STRUCTURAL BASIS OF ASYMMETRY IN THE HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 REVERSE TRANSCRIPTASE HETERODIMER | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P51), HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P66) | | Authors: | Steitz, T.A, Smerdon, S.J, Jaeger, J, Wang, J, Kohlstaedt, L.A, Chirino, A.J, Friedman, J.M, Rice, P.A. | | Deposit date: | 1994-07-25 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the binding site for nonnucleoside inhibitors of the reverse transcriptase of human immunodeficiency virus type 1.
Proc.Natl.Acad.Sci.Usa, 91, 1994
|
|
