6RKC
 
 | | Inter-dimeric interface controls function and stability of S-methionine adenosyltransferase from U. urealiticum | | Descriptor: | (DIPHOSPHONO)AMINOPHOSPHONIC ACID, MAGNESIUM ION, Methionine adenosyltransferase, ... | | Authors: | Shahar, A, Zarivach, R, Bershtein, S, Kleiner, D, Shmulevich, F. | | Deposit date: | 2019-04-30 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The interdimeric interface controls function and stability of Ureaplasma urealiticum methionine S-adenosyltransferase.
J.Mol.Biol., 431, 2019
|
|
1X9N
 
 | | Crystal Structure of Human DNA Ligase I bound to 5'-adenylated, nicked DNA | | Descriptor: | 5'-phosphorylated DNA, ADENOSINE MONOPHOSPHATE, DNA ligase I, ... | | Authors: | Pascal, J.M, O'Brien, P.J, Tomkinson, A.E, Ellenberger, T. | | Deposit date: | 2004-08-23 | | Release date: | 2004-11-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human DNA ligase I completely encircles and partially unwinds nicked DNA.
Nature, 432, 2004
|
|
6XR4
 
 | |
6RJS
 
 | | Inter-dimeric interface controls function and stability of S-methionine adenosyltransferase from U. urealiticum | | Descriptor: | Methionine adenosyltransferase | | Authors: | Shahar, A, Zarivach, R, Bershtein, S, Kleiner, D, Shmulevich, F. | | Deposit date: | 2019-04-29 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The interdimeric interface controls function and stability of Ureaplasma urealiticum methionine S-adenosyltransferase.
J.Mol.Biol., 431, 2019
|
|
3L2P
 
 | | Human DNA Ligase III Recognizes DNA Ends by Dynamic Switching Between Two DNA Bound States | | Descriptor: | 5'-D(*GP*CP*CP*AP*GP*TP*CP*CP*GP*AP*CP*GP*AP*CP*GP*CP*AP*TP*CP*CP*CP*G)-3', 5'-D(*GP*TP*CP*GP*GP*AP*CP*TP*G)-3', 5'-D(P*CP*GP*GP*GP*AP*TP*GP*CP*GP*TP*C)-3', ... | | Authors: | Cotner-Gohara, E.A, Kim, I.K, Hammel, M, Tainer, J.A, Tomkinson, A, Ellenberger, T. | | Deposit date: | 2009-12-15 | | Release date: | 2010-07-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human DNA Ligase III Recognizes DNA Ends by Dynamic Switching between Two DNA-Bound States.
Biochemistry, 49, 2010
|
|
7NH4
 
 | |
7NH5
 
 | |
3SO4
 
 | |
6S9W
 
 | |
6S9X
 
 | | Crystal Structure of AKT1 in Complex with Covalent-Allosteric AKT Inhibitor 15c | | Descriptor: | RAC-alpha serine/threonine-protein kinase, ~{N}-[3-[1-[[4-[5-[(4-hydroxyphenyl)methyl]-6-oxidanylidene-2-phenyl-1~{H}-pyrazin-3-yl]phenyl]methyl]piperidin-4-yl]-2-oxidanylidene-1~{H}-benzimidazol-5-yl]propanamide | | Authors: | Landel, I, Mueller, M.P, Rauh, D. | | Deposit date: | 2019-07-15 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Covalent-Allosteric Inhibitors to Achieve Akt Isoform-Selectivity.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6G52
 
 | | CRYSTAL STRUCTURE OF THE CNMP BINDING DOMAIN OF THE MAGNESIUM TRANSPORTER CNNM4 | | Descriptor: | Metal transporter CNNM4 | | Authors: | Gimenez, P, Oyenarte, I, Hardy, S, Zubillaga, M, Merino, N, Blanco, F.J, Siliqi, D, Tremblay, M, Muller, D, Martinez-Cruz, L.A. | | Deposit date: | 2018-03-28 | | Release date: | 2019-04-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.691 Å) | | Cite: | Structural Insights into the Intracellular Region of the Human Magnesium Transport Mediator CNNM4.
Int J Mol Sci, 20, 2019
|
|
8TYQ
 
 | | Structure of the C-terminal half of LRRK2 bound to GZD-824 (G2019S mutant) | | Descriptor: | 4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}-3-[(1H-pyrazolo[3,4-b]pyridin-5-yl)ethynyl]benzamide, Designed Ankyrin Repeats Protein E11, Leucine-rich repeat serine/threonine-protein kinase 2 | | Authors: | Villagran-Suarez, A, Sanz-Murillo, M, Alegrio-Louro, J, Leschziner, A. | | Deposit date: | 2023-08-25 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Inhibition of Parkinson's disease-related LRRK2 by type I and type II kinase inhibitors: Activity and structures.
Sci Adv, 9, 2023
|
|
8TZE
 
 | | Structure of C-terminal half of LRRK2 bound to GZD-824 | | Descriptor: | 4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}-3-[(1H-pyrazolo[3,4-b]pyridin-5-yl)ethynyl]benzamide, Leucine-rich repeat serine/threonine-protein kinase 2 | | Authors: | Villagran-Suarez, A, Sanz-Murillo, M, Alegrio-Louro, J, Leschziner, A. | | Deposit date: | 2023-08-26 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Inhibition of Parkinson's disease-related LRRK2 by type I and type II kinase inhibitors: Activity and structures.
Sci Adv, 9, 2023
|
|
8TZF
 
 | | Structure of full length LRRK2 bound to GZD-824 (I2020T mutant) | | Descriptor: | 4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}-3-[(1H-pyrazolo[3,4-b]pyridin-5-yl)ethynyl]benzamide, GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2, ... | | Authors: | Villagran-Suarez, A, Sanz-Murillo, M, Alegrio-Louro, J, Leschziner, A. | | Deposit date: | 2023-08-26 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Inhibition of Parkinson's disease-related LRRK2 by type I and type II kinase inhibitors: Activity and structures.
Sci Adv, 9, 2023
|
|
8TZH
 
 | | Structure of full-length LRRK2 bound to MLi-2 (I2020T mutant) | | Descriptor: | (2~{R},6~{S})-2,6-dimethyl-4-[6-[5-(1-methylcyclopropyl)oxy-1~{H}-indazol-3-yl]pyrimidin-4-yl]morpholine, E11 DARPin, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Sanz-Murillo, M, Villagran-Suarez, A, Alegrio Louro, J, Leschziner, A. | | Deposit date: | 2023-08-26 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Inhibition of Parkinson's disease-related LRRK2 by type I and type II kinase inhibitors: Activity and structures.
Sci Adv, 9, 2023
|
|
8TZB
 
 | | Structure of the C-terminal half of LRRK2 bound to GZD-824 (I2020T mutant) | | Descriptor: | 4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}-3-[(1H-pyrazolo[3,4-b]pyridin-5-yl)ethynyl]benzamide, Leucine-rich repeat serine/threonine-protein kinase 2, designed ankyrin repeat proteins E11 | | Authors: | Villagran-Suarez, A, Sanz-Murillo, M, Alegrio-Louro, J, Leschziner, A. | | Deposit date: | 2023-08-26 | | Release date: | 2023-12-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Inhibition of Parkinson's disease-related LRRK2 by type I and type II kinase inhibitors: Activity and structures.
Sci Adv, 9, 2023
|
|
8VDN
 
 | | DNA Ligase 1 with nick dG:C | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA ligase 1, Downstream Oligo, ... | | Authors: | KanalElamparithi, B, Gulkis, M, Caglayan, M. | | Deposit date: | 2023-12-16 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structures of LIG1 provide a mechanistic basis for understanding a lack of sugar discrimination against a ribonucleotide at the 3'-end of nick DNA.
J.Biol.Chem., 300, 2024
|
|
8VDS
 
 | | DNA Ligase 1 with nick DNA 3'rG:C | | Descriptor: | DNA (5'-D(*GP*TP*CP*CP*GP*AP*CP*CP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA ligase 1, DNA/RNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*T)-R(P*G)-D(P*GP*TP*CP*GP*GP*AP*C)-3') | | Authors: | KanalElamparithi, B, Gulkis, M, Caglayan, M. | | Deposit date: | 2023-12-17 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structures of LIG1 provide a mechanistic basis for understanding a lack of sugar discrimination against a ribonucleotide at the 3'-end of nick DNA.
J.Biol.Chem., 300, 2024
|
|
8VZM
 
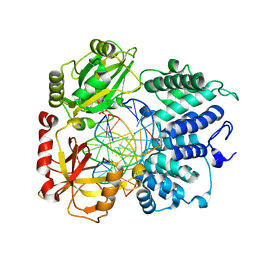 | | DNA Ligase 1 captured with pre-step 3 ligation at the rA:T nicksite | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA (5'-D(*GP*TP*CP*CP*GP*AP*CP*CP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(P*GP*TP*CP*GP*GP*AP*C)-3'), ... | | Authors: | KanalElamparithi, B, Gulkis, M, Caglayan, M. | | Deposit date: | 2024-02-11 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structures of LIG1 provide a mechanistic basis for understanding a lack of sugar discrimination against a ribonucleotide at the 3'-end of nick DNA.
J.Biol.Chem., 300, 2024
|
|
8VDT
 
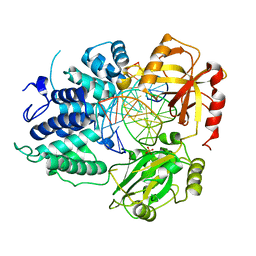 | | DNA Ligase 1 with nick DNA 3'rA:T | | Descriptor: | DNA (5'-D(*GP*TP*CP*CP*GP*AP*CP*TP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA ligase 1, DNA/RNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*T)-R(P*A)-D(P*GP*TP*CP*GP*GP*AP*C)-3'), ... | | Authors: | KanalElamparithi, B, Gulkis, M, Caglayan, M. | | Deposit date: | 2023-12-17 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structures of LIG1 provide a mechanistic basis for understanding a lack of sugar discrimination against a ribonucleotide at the 3'-end of nick DNA.
J.Biol.Chem., 300, 2024
|
|
8VZL
 
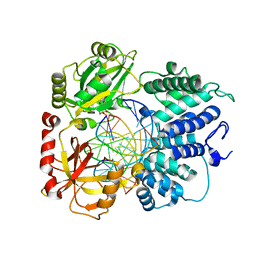 | | DNA Ligase 1 captured with pre-step 3 ligation at the rG:C nicksite | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA (5'-D(*GP*TP*CP*CP*GP*AP*CP*CP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(P*GP*TP*CP*GP*GP*AP*C)-3'), ... | | Authors: | KanalElamparithi, B, Gulkis, M, Caglayan, M. | | Deposit date: | 2024-02-11 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structures of LIG1 provide a mechanistic basis for understanding a lack of sugar discrimination against a ribonucleotide at the 3'-end of nick DNA.
J.Biol.Chem., 300, 2024
|
|
8BOT
 
 | |
8EOB
 
 | | Cryo-EM structure of human HSP90B in the closed state | | Descriptor: | Heat shock protein HSP 90-beta, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Srivastava, D, Artemyev, N.O. | | Deposit date: | 2022-10-02 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Unique interface and dynamics of the complex of HSP90 with a specialized cochaperone AIPL1.
Structure, 31, 2023
|
|
8EOA
 
 | | Cryo-EM structure of human HSP90B-AIPL1 complex | | Descriptor: | Aryl-hydrocarbon-interacting protein-like 1, Heat shock protein HSP 90-beta, MAGNESIUM ION, ... | | Authors: | Srivastava, D, Artemyev, N.O. | | Deposit date: | 2022-10-02 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Unique interface and dynamics of the complex of HSP90 with a specialized cochaperone AIPL1.
Structure, 31, 2023
|
|
8DBE
 
 | | Human PRPS1 with ADP; Hexamer | | Descriptor: | 5-O-phosphono-alpha-D-ribofuranose, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Hvorecny, K.L, Kollman, J.M. | | Deposit date: | 2022-06-14 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Human PRPS1 filaments stabilize allosteric sites to regulate activity.
Nat.Struct.Mol.Biol., 30, 2023
|
|
