3DZU
 
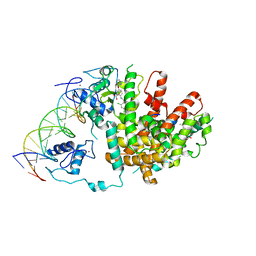 | | Intact PPAR gamma - RXR alpha Nuclear Receptor Complex on DNA bound with BVT.13, 9-cis Retinoic Acid and NCOA2 Peptide | | Descriptor: | (9cis)-retinoic acid, 2-[(2,4-DICHLOROBENZOYL)AMINO]-5-(PYRIMIDIN-2-YLOXY)BENZOIC ACID, DNA (5'-D(*DCP*DAP*DAP*DAP*DCP*DTP*DAP*DGP*DGP*DTP*DCP*DAP*DAP*DAP*DGP*DGP*DTP*DCP*DAP*DG)-3'), ... | | Authors: | Chandra, V, Huang, P, Hamuro, Y, Raghuram, S, Wang, Y, Burris, T.P, Rastinejad, F. | | Deposit date: | 2008-07-30 | | Release date: | 2008-10-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of the intact PPAR-gamma-RXR- nuclear receptor complex on DNA.
Nature, 456, 2008
|
|
3E00
 
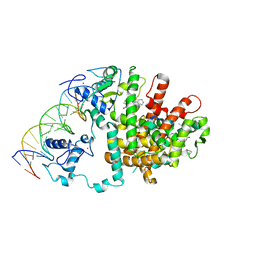 | | Intact PPAR gamma - RXR alpha Nuclear Receptor Complex on DNA bound with GW9662, 9-cis Retinoic Acid and NCOA2 Peptide | | Descriptor: | (9cis)-retinoic acid, 2-chloro-5-nitro-N-phenylbenzamide, DNA (5'-D(*DCP*DAP*DAP*DAP*DCP*DTP*DAP*DGP*DGP*DTP*DCP*DAP*DAP*DAP*DGP*DGP*DTP*DCP*DAP*DG)-3'), ... | | Authors: | Chandra, V, Huang, P, Hamuro, Y, Raghuram, S, Wang, Y, Burris, T.P, Rastinejad, F. | | Deposit date: | 2008-07-30 | | Release date: | 2008-10-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the intact PPAR-gamma-RXR- nuclear receptor complex on DNA.
Nature, 456, 2008
|
|
6CYT
 
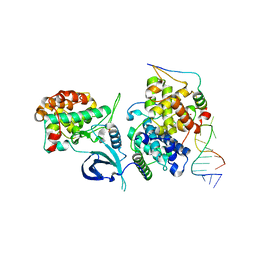 | | HIV-1 TAR loop in complex with Tat:AFF4:P-TEFb | | Descriptor: | AF4/FMR2 family member 4, Cyclin-T1, Cyclin-dependent kinase 9, ... | | Authors: | Schulze Gahmen, U, Hurley, J.H. | | Deposit date: | 2018-04-06 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural mechanism for HIV-1 TAR loop recognition by Tat and the super elongation complex.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5L1Z
 
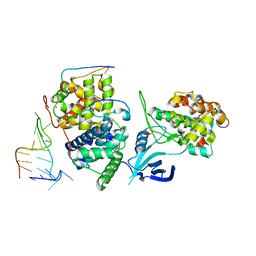 | | TAR complex with HIV-1 Tat-AFF4-P-TEFb | | Descriptor: | AF4/FMR2 family member 4, Cyclin-T1, Cyclin-dependent kinase 9, ... | | Authors: | Schulze-Gahmen, U, Hurley, J. | | Deposit date: | 2016-07-29 | | Release date: | 2016-10-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (5.9 Å) | | Cite: | Insights into HIV-1 proviral transcription from integrative structure and dynamics of the Tat:AFF4:P-TEFb:TAR complex.
Elife, 5, 2016
|
|
6BOS
 
 | |
6BOU
 
 | |
3GAT
 
 | | SOLUTION NMR STRUCTURE OF THE C-TERMINAL DOMAIN OF CHICKEN GATA-1 BOUND TO DNA, 34 STRUCTURES | | Descriptor: | DNA (5'-D(*AP*AP*TP*GP*TP*TP*TP*AP*TP*CP*TP*GP*CP*AP*AP*C)-3'), DNA (5'-D(*GP*TP*TP*GP*CP*AP*GP*AP*TP*AP*AP*AP*CP*AP*TP*T)-3'), ERYTHROID TRANSCRIPTION FACTOR GATA-1, ... | | Authors: | Clore, G.M, Tjandra, N, Starich, M, Omichinski, J.G, Gronenborn, A.M. | | Deposit date: | 1997-11-07 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Use of dipolar 1H-15N and 1H-13C couplings in the structure determination of magnetically oriented macromolecules in solution.
Nat.Struct.Biol., 4, 1997
|
|
2RAM
 
 | | A NOVEL DNA RECOGNITION MODE BY NF-KB P65 HOMODIMER | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, DNA (5'-D(*CP*GP*GP*CP*TP*GP*GP*AP*AP*AP*TP*(5IU)P*(5IU)P*CP*CP*AP*GP*CP*CP*G)-3'), PROTEIN (TRANSCRIPTION FACTOR NF-KB P65) | | Authors: | Chen, Y.Q, Ghosh, S, Ghosh, G. | | Deposit date: | 1997-11-23 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A novel DNA recognition mode by the NF-kappa B p65 homodimer.
Nat.Struct.Biol., 5, 1998
|
|
5U01
 
 | | Cooperative DNA binding by two RelA dimers | | Descriptor: | DNA (27-MER), Transcription factor p65 | | Authors: | Ghosh, G, Huang, D. | | Deposit date: | 2016-11-22 | | Release date: | 2017-09-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | DNA-binding affinity and transcriptional activity of the RelA homodimer of nuclear factor kappa B are not correlated.
J. Biol. Chem., 292, 2017
|
|
5UAN
 
 | | Crystal structure of multi-domain RAR-beta-RXR-alpha heterodimer on DNA | | Descriptor: | (9cis)-retinoic acid, DNA (5'-D(*CP*TP*AP*GP*GP*TP*CP*AP*AP*AP*GP*GP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*CP*CP*TP*TP*TP*GP*AP*CP*CP*TP*AP*G)-3'), ... | | Authors: | Chandra, V, Wu, D, Kim, Y, Rastinejad, F. | | Deposit date: | 2016-12-19 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.508 Å) | | Cite: | The quaternary architecture of RAR beta-RXR alpha heterodimer facilitates domain-domain signal transmission.
Nat Commun, 8, 2017
|
|
5SVZ
 
 | |
5V61
 
 | | Phospho-ERK2 bound to bivalent inhibitor SBP2 | | Descriptor: | 2-oxo-6,9,12,15-tetraoxa-3-azaoctadecan-18-oic acid, 5-(2-PHENYLPYRAZOLO[1,5-A]PYRIDIN-3-YL)-1H-PYRAZOLO[3,4-C]PYRIDAZIN-3-AMINE, GLYCEROL, ... | | Authors: | Lechtenberg, B.C, Riedl, S.J. | | Deposit date: | 2017-03-15 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Strategy for the Development of Potent Bivalent ERK Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
2WP2
 
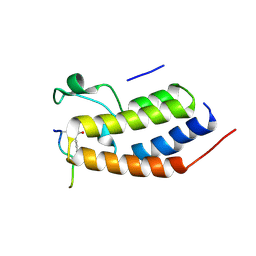 | | Structure of Brdt bromodomain BD1 bound to a diacetylated histone H4 peptide. | | Descriptor: | BROMODOMAIN TESTIS-SPECIFIC PROTEIN, HISTONE H4 | | Authors: | Moriniere, J, Rousseaux, S, Steuerwald, U, Soler-Lopez, M, Curtet, S, Vitte, A.-L, Govin, J, Gaucher, J, Sadoul, K, Hart, D.J, Krijgsveld, J, Khochbin, S, Mueller, C.W, Petosa, C. | | Deposit date: | 2009-08-02 | | Release date: | 2009-09-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Cooperative Binding of Two Acetylation Marks on a Histone Tail by a Single Bromodomain.
Nature, 461, 2009
|
|
2WP1
 
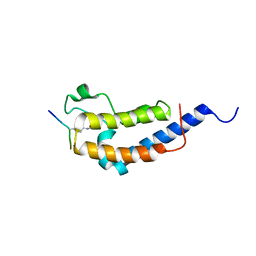 | | Structure of Brdt bromodomain 2 bound to an acetylated histone H3 peptide | | Descriptor: | BROMODOMAIN TESTIS-SPECIFIC PROTEIN, HISTONE H3 | | Authors: | Moriniere, J, Rousseaux, S, Steuerwald, U, Soler-Lopez, M, Curtet, S, Vitte, A.-L, Govin, J, Gaucher, J, Sadoul, K, Hart, D.J, Krijgsveld, J, Khochbin, S, Mueller, C.W, Petosa, C. | | Deposit date: | 2009-08-02 | | Release date: | 2009-09-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cooperative Binding of Two Acetylation Marks on a Histone Tail by a Single Bromodomain.
Nature, 461, 2009
|
|
6UV4
 
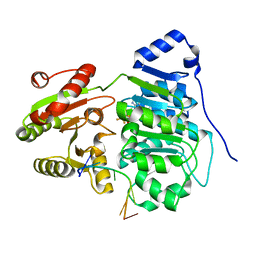 | | Crystal structure of the core domain of RNA helicase DDX17 with RNA pri-18a-oligo1 | | Descriptor: | 18a_oligo1, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Ngo, T.D, Partin, A.C, Nam, Y. | | Deposit date: | 2019-11-01 | | Release date: | 2020-01-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | RNA Specificity and Autoregulation of DDX17, a Modulator of MicroRNA Biogenesis.
Cell Rep, 29, 2019
|
|
6UV3
 
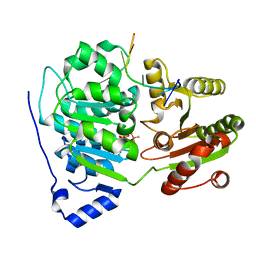 | | Crystal structure of the core domain of RNA helicase DDX17 with RNA pri-125a-oligo2 | | Descriptor: | 125a-oligo2, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Ngo, T.D, Partin, A.C, Nam, Y. | | Deposit date: | 2019-11-01 | | Release date: | 2020-01-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | RNA Specificity and Autoregulation of DDX17, a Modulator of MicroRNA Biogenesis.
Cell Rep, 29, 2019
|
|
6UV0
 
 | |
6UV1
 
 | | Crystal structure of RNA helicase DDX17 in complex of rU10 RNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Ngo, T.D, Partin, A.C, Nam, Y. | | Deposit date: | 2019-11-01 | | Release date: | 2020-01-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.307 Å) | | Cite: | RNA Specificity and Autoregulation of DDX17, a Modulator of MicroRNA Biogenesis.
Cell Rep, 29, 2019
|
|
4BCJ
 
 | | Structure of CDK9 in complex with cyclin T and a 2-amino-4-heteroaryl- pyrimidine inhibitor | | Descriptor: | 2-[(3-hydroxyphenyl)amino]-4-[4-methyl-2-(methylamino)-1,3-thiazol-5-yl]pyrimidine-5-carbonitrile, CYCLIN-DEPENDENT KINASE 9, CYCLIN-T1 | | Authors: | Hole, A.J, Baumli, S, Wang, S, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2012-10-02 | | Release date: | 2013-01-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.162 Å) | | Cite: | Comparative Structural and Functional Studies of 4-(Thiazol- 5-Yl)-2-(Phenylamino)Pyrimidine-5-Carbonitrile Cdk9 Inhibitors Suggest the Basis for Isotype Selectivity.
J.Med.Chem., 56, 2013
|
|
6UV2
 
 | | Crystal structure of the core domain of RNA helicase DDX17 with RNA pri-125a-oligo1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Ngo, T.D, Partin, A.C, Nam, Y. | | Deposit date: | 2019-11-01 | | Release date: | 2020-01-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.894 Å) | | Cite: | RNA Specificity and Autoregulation of DDX17, a Modulator of MicroRNA Biogenesis.
Cell Rep, 29, 2019
|
|
4BCG
 
 | | Structure of CDK9 in complex with cyclin T and a 2-amino-4-heteroaryl- pyrimidine inhibitor | | Descriptor: | 2-[[3-(1,4-diazepan-1-yl)phenyl]amino]-4-[4-methyl-2-(methylamino)-1,3-thiazol-5-yl]pyrimidine-5-carbonitrile, CYCLIN-DEPENDENT KINASE 9, CYCLIN-T1, ... | | Authors: | Hole, A.J, Baumli, S, Wang, S, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2012-10-02 | | Release date: | 2013-04-17 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (3.085 Å) | | Cite: | Substituted 4-(Thiazol-5-Yl)-2-(Phenylamino)Pyrimidines are Highly Active Cdk9 Inhibitors: Synthesis, X-Ray Crystal Structure, Sar and Anti-Cancer Activities.
J.Med.Chem., 56, 2013
|
|
7U50
 
 | |
6W9E
 
 | | Crystal Structure of Human CDK9/cyclinT1 in complex with MC180295 | | Descriptor: | (4-amino-2-{[(1R,2R,4S)-bicyclo[2.2.1]heptan-2-yl]amino}-1,3-thiazol-5-yl)(2-nitrophenyl)methanone, (4-amino-2-{[(1S,2S,4R)-bicyclo[2.2.1]heptan-2-yl]amino}-1,3-thiazol-5-yl)(2-nitrophenyl)methanone, Cyclin-T1, ... | | Authors: | Zhang, P, Wu, J. | | Deposit date: | 2020-03-22 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Comparative Modeling of CDK9 Inhibitors to Explore Selectivity and Structure-Activity Relationships
To Be Published
|
|
4BCI
 
 | | Structure of CDK9 in complex with cyclin T and a 2-amino-4-heteroaryl- pyrimidine inhibitor | | Descriptor: | 3-[[5-cyano-4-[4-methyl-2-(methylamino)-1,3-thiazol-5-yl]pyrimidin-2-yl]amino]benzenesulfonamide, CYCLIN-DEPENDENT KINASE 9, CYCLIN-T1 | | Authors: | Hole, A.J, Baumli, S, Wang, S, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2012-10-02 | | Release date: | 2013-01-09 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Comparative Structural and Functional Studies of 4-(Thiazol- 5-Yl)-2-(Phenylamino)Pyrimidine-5-Carbonitrile Cdk9 Inhibitors Suggest the Basis for Isotype Selectivity.
J.Med.Chem., 56, 2013
|
|
4BCH
 
 | | Structure of CDK9 in complex with cyclin T and a 2-amino-4-heteroaryl- pyrimidine inhibitor | | Descriptor: | 4-(4-methyl-2-methylimino-3H-1,3-thiazol-5-yl)-2-[(4-methyl-3-morpholin-4-ylsulfonyl-phenyl)amino]pyrimidine-5-carbonitrile, CYCLIN-DEPENDENT KINASE 9, CYCLIN-T1, ... | | Authors: | Hole, A.J, Baumli, S, Wang, S, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2012-10-02 | | Release date: | 2013-01-09 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (2.958 Å) | | Cite: | Comparative Structural and Functional Studies of 4-(Thiazol- 5-Yl)-2-(Phenylamino)Pyrimidine-5-Carbonitrile Cdk9 Inhibitors Suggest the Basis for Isotype Selectivity.
J.Med.Chem., 56, 2013
|
|
