6DUC
 
 | | Crystal structure of mutant beta-K167T tryptophan synthase in complex with inhibitor N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the alpha-site, cesium ion at the metal coordination site, and 2-aminophenol quinonoid (1D0) at the beta-site | | Descriptor: | (2E)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]-3-[(2-hydroxyphenyl)amino]propanoic acid, 1,2-ETHANEDIOL, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, ... | | Authors: | Hilario, E, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2018-06-20 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Tryptophan synthase Q114A mutant in complex with inhibitor N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the alpha-site, aminoacrylate (P1T) at the beta site, and cesium ion at the metal coordination site.
To be Published
|
|
2BZE
 
 | | NMR Structure of human RTF1 PLUS3 domain. | | Descriptor: | KIAA0252 PROTEIN | | Authors: | Truffault, V, Diercks, T, Ab, E, De Jong, R.N, Daniels, M.A, Kaptein, R, Folkers, G.E, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-08-16 | | Release date: | 2007-01-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure and DNA Binding of the Human Rtf1 Plus3 Domain.
Structure, 16, 2008
|
|
6I23
 
 | | Flavin Analogue Sheds Light on Light-Oxygen-Voltage Domain Mechanism | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-1-(7,8-dimethyl-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-5-O-phosphono-D-ribitol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Rizkallah, P.J, Kalvaitis, M.E, Allemann, R.K, Mart, R.J, Johnson, L.A. | | Deposit date: | 2018-10-31 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Noncanonical Chromophore Reveals Structural Rearrangements of the Light-Oxygen-Voltage Domain upon Photoactivation.
Biochemistry, 58, 2019
|
|
8PCW
 
 | | Structure of Csm6' from Streptococcus thermophilus | | Descriptor: | CRISPR system endoribonuclease Csm6' | | Authors: | McQuarrie, S.J, Athukoralage, J.S, McMahon, S.A, Graham, S, Ackerman, K, Bode, B.E, White, M.F, Gloster, T.M. | | Deposit date: | 2023-06-11 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.54 Å) | | Cite: | Activation of Csm6 ribonuclease by cyclic nucleotide binding: in an emergency, twist to open.
Nucleic Acids Res., 51, 2023
|
|
2YPP
 
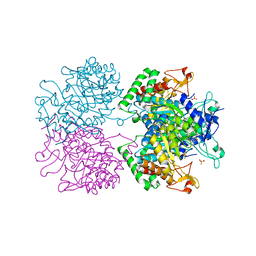 | | 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase in complex with 3 tyrosine molecules | | Descriptor: | CHLORIDE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Blackmore, N.J, Reichau, S, Jiao, W, Hutton, R.D, Baker, E.N, Jameson, G.B, Parker, E.J. | | Deposit date: | 2012-10-31 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three Sites and You are Out: Ternary Synergistic Allostery Controls Aromatic Aminoacid Biosynthesis in Mycobacterium Tuberculosis.
J.Mol.Biol., 425, 2013
|
|
1SCF
 
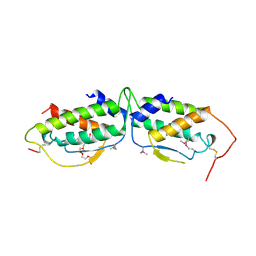 | | HUMAN RECOMBINANT STEM CELL FACTOR | | Descriptor: | CALCIUM ION, PENTAETHYLENE GLYCOL, STEM CELL FACTOR | | Authors: | Jiang, X, Gurel, O, Langley, K.E, Hendrickson, W.A. | | Deposit date: | 1998-06-04 | | Release date: | 2000-07-07 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the active core of human stem cell factor and analysis of binding to its receptor kit.
EMBO J., 19, 2000
|
|
3LVM
 
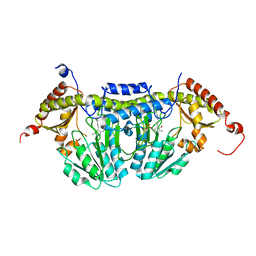 | | Crystal Structure of E.coli IscS | | Descriptor: | Cysteine desulfurase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Shi, R, Proteau, A, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-02-22 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for Fe-S cluster assembly and tRNA thiolation mediated by IscS protein-protein interactions.
Plos Biol., 8, 2010
|
|
4BV7
 
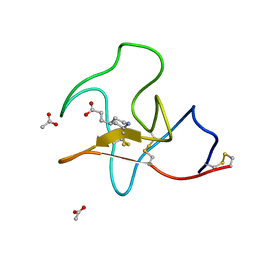 | | Identification of small molecule inhibitors selective for apo(a) kringles KIV-7, KIV-10 and KV. | | Descriptor: | 3-(4-piperidyl)propanoic acid, ACETATE ION, APOLIPOPROTEIN(A) | | Authors: | Sandmark, J, Althage, M, Andersson, G.M.K, Antonsson, T, Blaho, S, Bodin, C, Bostrom, J, Chen, Y, Dahlen, A, Eriksson, P.O, Evertsson, E, Fex, T, Fjellstrom, O, Gustafsson, D, Hallberg, C, Hicks, R, Jarkvist, E, Johansson, C, Kalies, I, Kang, D, Svalstedt Karlsson, B, Kartberg, F, Legnehed, A, Lindqvist, A.M, Martinsson, S.A, Moberg, A, Petersson, A.U, Ridderstrom, M, Thelin, A, Tigerstrom, A, Vinblad, J, Xu, B, Knecht, W. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Small Molecules Used to Decipher the Pathophysiological Roles of the Kringle Domains Kiv-7, - 10 and Kv of Apolipoprotein(A)
To be Published
|
|
2LQX
 
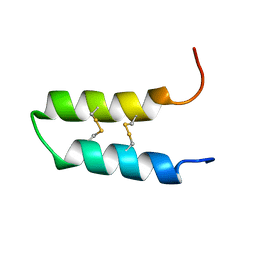 | | NMR spatial structure of the trypsin inhibitor BWI-2c from the buckwheat seeds | | Descriptor: | Trypsin inhibitor BWI-2c | | Authors: | Mineev, K.S, Vassilevski, A.A, Oparin, P.B, Grishin, E.V, Egorov, T.A. | | Deposit date: | 2012-03-19 | | Release date: | 2012-05-30 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Buckwheat trypsin inhibitor with helical hairpin structure belongs to a new family of plant defence peptides.
Biochem.J., 446, 2012
|
|
1SFQ
 
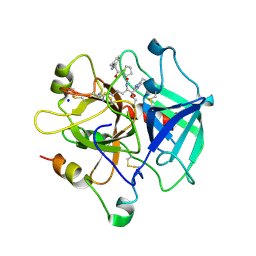 | | Fast form of thrombin mutant R(77a)A bound to PPACK | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, SODIUM ION, ... | | Authors: | Pineda, A.O, Carrell, C.J, Bush, L.A, Prasad, S, Caccia, S, Chen, Z.W, Mathews, F.S, Di Cera, E. | | Deposit date: | 2004-02-20 | | Release date: | 2004-06-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Molecular dissection of na+ binding to thrombin.
J.Biol.Chem., 279, 2004
|
|
2HGT
 
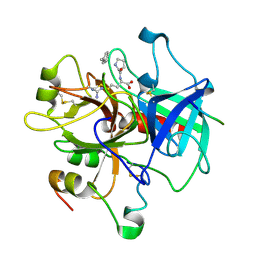 | |
2C0S
 
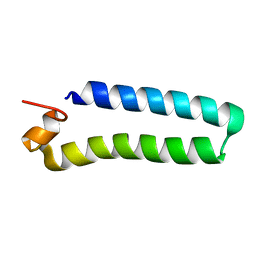 | | NMR Solution Structure of a protein aspartic acid phosphate phosphatase from Bacillus Anthracis | | Descriptor: | CONSERVED DOMAIN PROTEIN | | Authors: | Grenha, R, Rzechorzek, N.J, Brannigan, J.A, Ab, E, Folkers, G.E, De Jong, R.N, Diercks, T, Wilkinson, A.J, Kaptein, R, Wilson, K.S. | | Deposit date: | 2005-09-07 | | Release date: | 2006-09-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of Spo0E-like protein-aspartic acid phosphatases that regulate sporulation in bacilli.
J. Biol. Chem., 281, 2006
|
|
2F57
 
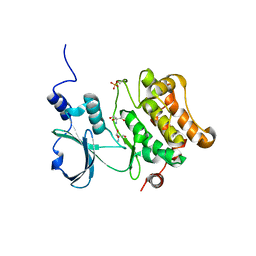 | | Crystal Structure Of The Human P21-Activated Kinase 5 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N2-[(1R,2S)-2-AMINOCYCLOHEXYL]-N6-(3-CHLOROPHENYL)-9-ETHYL-9H-PURINE-2,6-DIAMINE, Serine/threonine-protein kinase PAK 7 | | Authors: | Eswaran, J, Turnbull, A, Ugochukwu, E, Papagrigoriou, E, Bray, J, Das, S, Savitsky, P, Smee, C, Burgess, N, Fedorov, O, Filippakopoulos, P, von Delft, F, Arrowsmith, C, Weigelt, J, Sundstrom, M, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-11-25 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of the p21-activated kinases PAK4, PAK5, and PAK6 reveal catalytic domain plasticity of active group II PAKs.
Structure, 15, 2007
|
|
2C4F
 
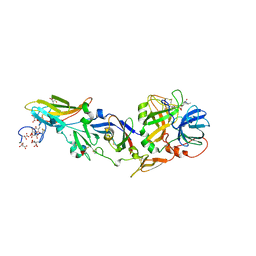 | | crystal structure of factor VII.stf complexed with pd0297121 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{[6-{3-[AMINO(IMINO)METHYL]PHENOXY}-4-(DIISOPROPYLAMINO)-3,5-DIFLUOROPYRIDIN-2-YL]OXY}-5-[(ISOBUTYLAMINO)CARBONYL]BEN ZOIC ACID, CALCIUM ION, ... | | Authors: | Kohrt, J.T, Zhang, E. | | Deposit date: | 2005-10-18 | | Release date: | 2006-10-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The Discovery of Fluoropyridine-Based Inhibitors of the Factor Viia/Tf Complex--Part 2
Bioorg.Med.Chem.Lett., 16, 2006
|
|
3H2V
 
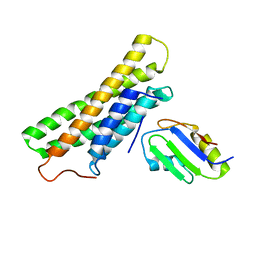 | | Human raver1 RRM1 domain in complex with human vinculin tail domain Vt | | Descriptor: | Raver-1, Vinculin | | Authors: | Lee, J.H, Rangarajan, E.S, Yogesha, S.D, Izard, T. | | Deposit date: | 2009-04-14 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Raver1 interactions with vinculin and RNA suggest a feed-forward pathway in directing mRNA to focal adhesions
Structure, 17, 2009
|
|
4BVC
 
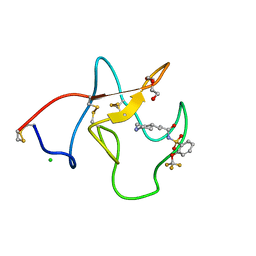 | | Identification of small molecule inhibitors selective for apo(a) kringles KIV-7, KIV-10 and KV. | | Descriptor: | 3-(4-PIPERIDYL)-N-[2-(TRIFLUOROMETHOXY)PHENYL]SULFONYL-PROPANAMIDE, APOLIPOPROTEIN(A), CHLORIDE ION, ... | | Authors: | Sandmark, J, Althage, M, Andersson, G.M.K, Antonsson, T, Blaho, S, Bodin, C, Bostrom, J, Chen, Y, Dahlen, A, Eriksson, P.O, Evertsson, E, Fex, T, Fjellstrom, O, Gustafsson, D, Hallberg, C, Hicks, R, Jarkvist, E, Johansson, C, Kalies, I, Kang, D, Svalstedt Karlsson, B, Kartberg, F, Legnehed, A, Lindqvist, A.M, Martinsson, S.A, Moberg, A, Petersson, A.U, Ridderstrom, M, Thelin, A, Tigerstrom, A, Vinblad, J, Xu, B, Knecht, W. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Small Molecules Used to Decipher the Pathophysiological Roles of the Kringle Domains Kiv-7, - 10 and Kv of Apolipoprotein(A)
To be Published
|
|
4KEL
 
 | | Atomic resolution crystal structure of Kallikrein-Related Peptidase 4 complexed with a modified SFTI inhibitor FCQR(N) | | Descriptor: | Kallikrein-4, Trypsin inhibitor 1 | | Authors: | Ilyichova, O.V, Swedberg, J.E, de Veer, S.J, Sit, K.C, Harris, J.M, Buckle, A.M. | | Deposit date: | 2013-04-25 | | Release date: | 2014-04-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.148 Å) | | Cite: | KLK4 Inhibition by Cyclic and Acyclic Peptides: Structural and Dynamical Insights into Standard-Mechanism Protease Inhibitors.
Biochemistry, 58, 2019
|
|
6E02
 
 | | WT swMb-MeNO | | Descriptor: | Myoglobin, NITROSOMETHANE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Herrera, V.E. | | Deposit date: | 2018-07-05 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Insights into Nitrosoalkane Binding to Myoglobin Provided by Crystallography of Wild-Type and Distal Pocket Mutant Derivatives.
Biochemistry, 62, 2023
|
|
8EVK
 
 | | Crystal structure of Helicobacter pylori dihydroneopterin aldolase (DHNA) | | Descriptor: | 1,2-ETHANEDIOL, Dihydroneopterin aldolase, PTERINE | | Authors: | Shaw, G.X, Cherry, S, Tropea, J.E, Ji, X. | | Deposit date: | 2022-10-20 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structure of Helicobacter pylori dihydroneopterin aldolase suggests a fragment-based strategy for isozyme-specific inhibitor design.
Curr Res Struct Biol, 5, 2023
|
|
6IA7
 
 | | T. brucei IFT22 GTP-bound crystal structure | | Descriptor: | CALCIUM ION, GUANOSINE-5'-TRIPHOSPHATE, Intraflagellar transport protein 22, ... | | Authors: | Wachter, S, Basquin, J, Lorentzen, E. | | Deposit date: | 2018-11-26 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Binding of IFT22 to the intraflagellar transport complex is essential for flagellum assembly.
Embo J., 38, 2019
|
|
6IAE
 
 | | T. brucei IFT22 GDP-bound crystal structure | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Intraflagellar transport protein 22, MAGNESIUM ION | | Authors: | Wachter, S, Basquin, J, Lorentzen, E. | | Deposit date: | 2018-11-26 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Binding of IFT22 to the intraflagellar transport complex is essential for flagellum assembly.
Embo J., 38, 2019
|
|
4ZNW
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in complex with a 4-Bromo-substituted OBHS derivative | | Descriptor: | 4-bromophenyl (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate, Estrogen receptor, Nuclear receptor-interacting peptide | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-05-05 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
3H70
 
 | | Crystal structure of o-succinylbenzoic acid synthetase from staphylococcus aureus Complexed with mg in the active site | | Descriptor: | MAGNESIUM ION, O-succinylbenzoic acid (OSB) synthetase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-24 | | Release date: | 2009-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4ZGK
 
 | | Structure of Mdm2 with low molecular weight inhibitor. | | Descriptor: | (5R)-3,5-bis(4-chlorobenzyl)-4-(6-chloro-1H-indol-3-yl)-5-hydroxyfuran-2(5H)-one, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Twarda-Clapa, A, Zak, K.M, Wrona, E.M, Grudnik, P, Dubin, G, Holak, T.A. | | Deposit date: | 2015-04-23 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Unique Mdm2-Binding Mode of the 3-Pyrrolin-2-one- and 2-Furanone-Based Antagonists of the p53-Mdm2 Interaction.
ACS Chem. Biol., 11, 2016
|
|
6E03
 
 | | sperm whale myoglobin nitrosoethane adduct | | Descriptor: | CHLORIDE ION, Myoglobin, NITROSOETHANE, ... | | Authors: | Herrera, V.E. | | Deposit date: | 2018-07-05 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Insights into Nitrosoalkane Binding to Myoglobin Provided by Crystallography of Wild-Type and Distal Pocket Mutant Derivatives.
Biochemistry, 62, 2023
|
|
