5VQF
 
 | | Crystal Structure of pro-TGF-beta 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Transforming growth factor beta-1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Zhao, B, Xu, S, Dong, X, Lu, C, Springer, T.A. | | Deposit date: | 2017-05-08 | | Release date: | 2017-11-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Prodomain-growth factor swapping in the structure of pro-TGF-beta 1.
J. Biol. Chem., 293, 2018
|
|
2BL8
 
 | | 1.6 Angstrom crystal structure of EntA-im: a bacterial immunity protein conferring immunity to the antimicrobial activity of the pediocin-like bacteriocin, enterocin A | | Descriptor: | CITRATE ANION, ENTEROCINE A IMMUNITY PROTEIN | | Authors: | Johnsen, L, Dalhus, B, Leiros, I, Nissen-Meyer, J. | | Deposit date: | 2005-03-02 | | Release date: | 2005-03-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 1.6-A Crystal Structure of Enta-Im: A Bacterial Immunity Protein Conferring Immunity to the Antimicrobial Activity of the Pediocin-Like Bacteriocin Enterocin A
J.Biol.Chem., 280, 2005
|
|
2BTE
 
 | | Thermus thermophilus Leucyl-tRNA synthetase complexed with a tRNAleu transcript in the post-editing conformation and a post- transfer editing substrate analogue | | Descriptor: | 2'-AMINO-2'-DEOXYADENOSINE, AMINOACYL-TRNA SYNTHETASE, LEUCINE, ... | | Authors: | Cusack, S, Tukalo, M, Yaremchuk, A, Fukunaga, R, Yokoyama, S. | | Deposit date: | 2005-05-31 | | Release date: | 2005-09-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Crystal Structure of Leucyl-tRNA Synthetase Complexed with tRNA(Leu) in the Post-Transfer- Editing Conformation.
Nat.Struct.Mol.Biol., 12, 2005
|
|
5VJI
 
 | | Crystal structure of the CLOCK Transcription Domain Exon19 in Complex with a Repressor | | Descriptor: | CLOCK-interacting pacemaker, Circadian locomoter output cycles protein kaput | | Authors: | Hou, Z, Su, L, Pei, J, Grishin, N.V, Zhang, H. | | Deposit date: | 2017-04-19 | | Release date: | 2017-06-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal Structure of the CLOCK Transactivation Domain Exon19 in Complex with a Repressor.
Structure, 25, 2017
|
|
6ZM7
 
 | | SARS-CoV-2 Nsp1 bound to the human CCDC124-80S-EBP1 ribosome complex | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-01 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
6ZME
 
 | | SARS-CoV-2 Nsp1 bound to the human CCDC124-80S-eERF1 ribosome complex | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-02 | | Release date: | 2020-08-12 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
6PFJ
 
 | | Structure of S. venezuelae RsiG-WhiG-(ci-di-GMP) complex, P64 crystal form | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), AmfC protein, RNA polymerase sigma factor | | Authors: | Schumacher, M.A. | | Deposit date: | 2019-06-21 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | c-di-GMP Arms an Anti-sigma to Control Progression of Multicellular Differentiation in Streptomyces.
Mol.Cell, 77, 2020
|
|
6PFV
 
 | | Structure of S. venezuelae RisG-WhiG-c-di-GMP complex: orthorhombic crystal form | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), AmfC protein, RNA polymerase sigma factor | | Authors: | Schumacher, M.A. | | Deposit date: | 2019-06-22 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | c-di-GMP Arms an Anti-sigma to Control Progression of Multicellular Differentiation in Streptomyces.
Mol.Cell, 77, 2020
|
|
5KCV
 
 | | Crystal structure of allosteric inhibitor, ARQ 092, in complex with autoinhibited form of AKT1 | | Descriptor: | 3-[3-[4-(1-azanylcyclobutyl)phenyl]-5-phenyl-imidazo[4,5-b]pyridin-2-yl]pyridin-2-amine, RAC-alpha serine/threonine-protein kinase | | Authors: | Eathiraj, S. | | Deposit date: | 2016-06-07 | | Release date: | 2016-06-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of 3-(3-(4-(1-Aminocyclobutyl)phenyl)-5-phenyl-3H-imidazo[4,5-b]pyridin-2-yl)pyridin-2-amine (ARQ 092): An Orally Bioavailable, Selective, and Potent Allosteric AKT Inhibitor.
J.Med.Chem., 59, 2016
|
|
7OIZ
 
 | | Cryo-EM structure of 70S ribosome stalled with TnaC peptide | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Su, T, Kudva, R, Becker, T, Berninghausen, O, Heijne, G, Cheng, J, Beckmann, R. | | Deposit date: | 2021-05-13 | | Release date: | 2021-09-15 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of l-tryptophan-dependent inhibition of release factor 2 by the TnaC arrest peptide.
Nucleic Acids Res., 49, 2021
|
|
5KPM
 
 | | Glycogen Synthase Kinase 3 beta Complexed with BRD3731 | | Descriptor: | (4~{S})-3-(2,2-dimethylpropyl)-4,7,7-trimethyl-4-phenyl-2,6,8,9-tetrahydropyrazolo[3,4-b]quinolin-5-one, Glycogen synthase kinase-3 beta | | Authors: | Lakshminarasimhan, D, White, A, Nadupalli, A, Suto, R.K. | | Deposit date: | 2016-07-04 | | Release date: | 2018-03-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Exploiting an Asp-Glu "switch" in glycogen synthase kinase 3 to design paralog-selective inhibitors for use in acute myeloid leukemia.
Sci Transl Med, 10, 2018
|
|
5KPL
 
 | | Glycogen Synthase Kinase 3 beta Complexed with BRD0705 | | Descriptor: | (4~{S})-4-ethyl-7,7-dimethyl-4-phenyl-2,6,8,9-tetrahydropyrazolo[3,4-b]quinolin-5-one, Glycogen synthase kinase-3 beta | | Authors: | Lakshminarasimhan, D, White, A, Nadupalli, A, Suto, R.K. | | Deposit date: | 2016-07-04 | | Release date: | 2018-03-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Exploiting an Asp-Glu "switch" in glycogen synthase kinase 3 to design paralog-selective inhibitors for use in acute myeloid leukemia.
Sci Transl Med, 10, 2018
|
|
5KPK
 
 | | Glycogen Synthase Kinase 3 beta Complexed with BRD0209 | | Descriptor: | (4~{S})-3-cyclopropyl-4,7,7-trimethyl-4-phenyl-2,6,8,9-tetrahydropyrazolo[3,4-b]quinolin-5-one, Glycogen synthase kinase-3 beta | | Authors: | Lakshminarasimhan, D, White, A, Nadupalli, A, Suto, R.K. | | Deposit date: | 2016-07-04 | | Release date: | 2018-03-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Exploiting an Asp-Glu "switch" in glycogen synthase kinase 3 to design paralog-selective inhibitors for use in acute myeloid leukemia.
Sci Transl Med, 10, 2018
|
|
4V5Q
 
 | | The crystal structure of EF-Tu and G24A-tRNA-Trp bound to a near- cognate codon on the 70S ribosome | | Descriptor: | 16S RRNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Schmeing, T.M, Voorhees, R.M, Ramakrishnan, V. | | Deposit date: | 2010-12-07 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | How Mutations in tRNA Distant from the Anticodon Affect the Fidelity of Decoding.
Nat.Struct.Mol.Biol., 18, 2011
|
|
4V90
 
 | | Thermus thermophilus Ribosome | | Descriptor: | 16S RIBOSOMAL RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Chen, Y, Feng, S, Kumar, V, Ero, R, Gao, Y.G. | | Deposit date: | 2014-02-22 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of EF-G-ribosome complex in a pretranslocation state.
Nat. Struct. Mol. Biol., 20, 2013
|
|
6KDH
 
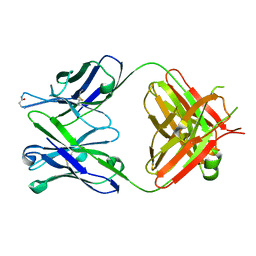 | | Antibody 64M-5 Fab including isoAsp in ligand-free form | | Descriptor: | Anti-(6-4) photoproduct antibody 64M-5 Fab (heavy chain), Anti-(6-4) photoproduct antibody 64M-5 Fab (light chain) | | Authors: | Yokoyama, H, Mizutani, R, Noguchi, S, Hayashida, N. | | Deposit date: | 2019-07-02 | | Release date: | 2019-12-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural and biochemical basis of the formation of isoaspartate in the complementarity-determining region of antibody 64M-5 Fab.
Sci Rep, 9, 2019
|
|
4V5R
 
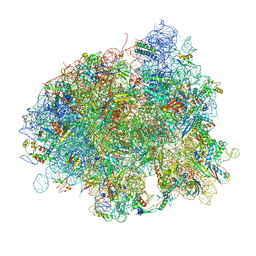 | | The crystal structure of EF-Tu and Trp-tRNA-Trp bound to a cognate codon on the 70S ribosome. | | Descriptor: | 16S RRNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Schmeing, T.M, Voorhees, R.M, Ramakrishnan, V. | | Deposit date: | 2010-12-07 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | How Mutations in tRNA Distant from the Anticodon Affect the Fidelity of Decoding.
Nat.Struct.Mol.Biol., 18, 2011
|
|
8K2C
 
 | | Cryo-EM structure of the human 80S ribosome with Tigecycline | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Li, X, Wang, M, Cheng, J. | | Deposit date: | 2023-07-12 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
8XSY
 
 | | Cryo-EM structure of the human 80S ribosome with Tigecycline, e-tRNA and CCDC124 (40S head Swivelled) | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Li, X, Wang, M, Cheng, J. | | Deposit date: | 2024-01-10 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
5DFE
 
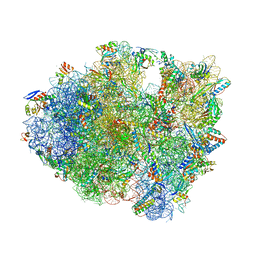 | | 70S termination complex containing E. coli RF2 | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Hoffer, E.D, Dunham, C.M. | | Deposit date: | 2015-08-26 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.09997559 Å) | | Cite: | Uniformity of Peptide Release Is Maintained by Methylation of Release Factors.
Cell Rep, 17, 2016
|
|
5D8B
 
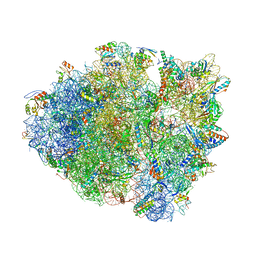 | |
1S1T
 
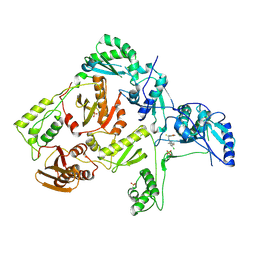 | | Crystal structure of L100I mutant HIV-1 reverse transcriptase in complex with UC-781 | | Descriptor: | 2-METHYL-FURAN-3-CARBOTHIOIC ACID [4-CHLORO-3-(3-METHYL-BUT-2-ENYLOXY)-PHENYL]-AMIDE, PHOSPHATE ION, Reverse transcriptase | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2004-01-07 | | Release date: | 2004-06-29 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptases mutated at codons 100, 106 and 108 and mechanisms of resistance to non-nucleoside inhibitors
J.Mol.Biol., 336, 2004
|
|
5DHY
 
 | | HIV-1 Rev NTD dimers with variable crossing angles | | Descriptor: | Anti-Rev Antibody Fab single-chain variable fragment, heavy chain, light chain, ... | | Authors: | DiMattia, M.A, Watts, N.R, Wingfield, P.T, Grimes, J.M, Stuart, D.I, Steven, A.C. | | Deposit date: | 2015-08-31 | | Release date: | 2016-06-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Structure of HIV-1 Rev Filaments Suggests a Bilateral Model for Rev-RRE Assembly.
Structure, 24, 2016
|
|
5DHX
 
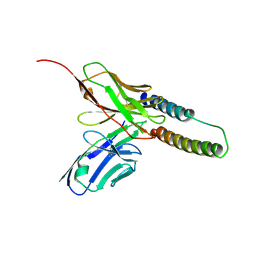 | | HIV-1 Rev NTD dimers with variable crossing angles | | Descriptor: | Anti-Rev Antibody Fab single-chain variable fragment, light chain,Anti-Rev Antibody Fab single-chain variable fragment, heavy chain, ... | | Authors: | DiMattia, M.A, Watts, N.R, Wingfield, P.T, Grimes, J.M, Stuart, D.I, Steven, A.C. | | Deposit date: | 2015-08-31 | | Release date: | 2016-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Structure of HIV-1 Rev Filaments Suggests a Bilateral Model for Rev-RRE Assembly.
Structure, 24, 2016
|
|
5DHZ
 
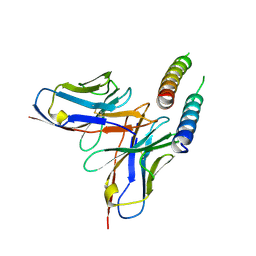 | | HIV-1 Rev NTD dimers with variable crossing angles | | Descriptor: | Anti-Rev Antibody Fab single-chain variable fragment, heavy chain, light chain, ... | | Authors: | DiMattia, M.A, Watts, N.R, Wingfield, P.T, Grimes, J.M, Stuart, D.I, Steven, A.C. | | Deposit date: | 2015-08-31 | | Release date: | 2016-06-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | The Structure of HIV-1 Rev Filaments Suggests a Bilateral Model for Rev-RRE Assembly.
Structure, 24, 2016
|
|
