7R1Z
 
 | |
2QPA
 
 | | Crystal Structure of S.cerevisiae Vps4 in the presence of ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, Vacuolar protein sorting-associated protein 4 | | Authors: | Xiao, J, Xu, Z. | | Deposit date: | 2007-07-23 | | Release date: | 2007-10-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural characterization of the ATPase reaction cycle of endosomal AAA protein Vps4.
J.Mol.Biol., 374, 2007
|
|
3SJ2
 
 | | A Crystal Structure of a Model of the Repeating r(CGG) Transcript Found in Fragile X Syndrome | | Descriptor: | ACETATE ION, RNA (5'-R(*UP*UP*GP*GP*GP*CP*CP*GP*GP*CP*GP*GP*CP*GP*GP*GP*UP*CP*C)-3'), RNA (5'-R(P*GP*GP*GP*CP*CP*GP*GP*CP*GP*GP*CP*GP*GP*GP*UP*CP*C)-3') | | Authors: | Kumar, A, Pengfei, F, Park, H, Nettles, K, Guo, M, Disney, M.D. | | Deposit date: | 2011-06-20 | | Release date: | 2011-08-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | A Crystal Structure of a Model of the Repeating r(CGG) Transcript Found in Fragile X Syndrome.
Chembiochem, 12, 2011
|
|
2QRV
 
 | | Structure of Dnmt3a-Dnmt3L C-terminal domain complex | | Descriptor: | DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3A, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Jia, D, Cheng, X. | | Deposit date: | 2007-07-29 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation.
Nature, 449, 2007
|
|
2QJC
 
 | | Crystal structure of a putative diadenosine tetraphosphatase | | Descriptor: | Diadenosine tetraphosphatase, putative, MANGANESE (II) ION, ... | | Authors: | Sugadev, R, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-07-06 | | Release date: | 2007-07-24 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
1TJL
 
 | | Crystal structure of transcription factor DksA from E. coli | | Descriptor: | DnaK suppressor protein, ZINC ION | | Authors: | Perederina, A, Svetlov, V, Vassylyeva, M.N, Artsimovitch, I, Yokoyama, S, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-06-06 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Regulation through the secondary channel--structural framework for ppGpp-DksA synergism during transcription
Cell(Cambridge,Mass.), 118, 2004
|
|
3C5G
 
 | | Structure of a ternary complex of the R517K Pol lambda mutant | | Descriptor: | 1,2-ETHANEDIOL, 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, DNA (5'-D(*DCP*DAP*DGP*DTP*DAP*(2DT))-3'), ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Foley, M.C, Pedersen, L.C, Schlick, T, Kunkel, T.A. | | Deposit date: | 2008-01-31 | | Release date: | 2008-07-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate-induced DNA strand misalignment during catalytic cycling by DNA polymerase lambda.
Embo Rep., 9, 2008
|
|
4XKP
 
 | |
6W7F
 
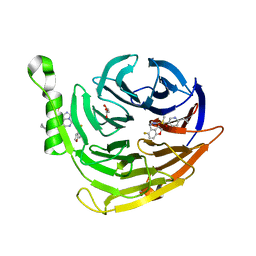 | | Structure of EED bound to inhibitor 5285 | | Descriptor: | 8-(6-cyclopropylpyridin-3-yl)-N-[(5-fluoro-2,3-dihydro-1-benzofuran-4-yl)methyl]-1-(methylsulfonyl)imidazo[1,5-c]pyrimidin-5-amine, GLYCEROL, Polycomb protein EED | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2020-03-19 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | EEDi-5285: An Exceptionally Potent, Efficacious, and Orally Active Small-Molecule Inhibitor of Embryonic Ectoderm Development.
J.Med.Chem., 63, 2020
|
|
6WBW
 
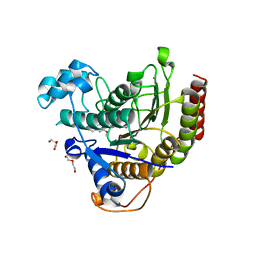 | | Structure of Human HDAC2 in complex with an ethyl ketone inhibitor | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Histone deacetylase 2, ... | | Authors: | Klein, D.J, Yu, W. | | Deposit date: | 2020-03-27 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Discovery of ethyl ketone-based HDACs 1, 2, and 3 selective inhibitors for HIV latency reactivation.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
2VJV
 
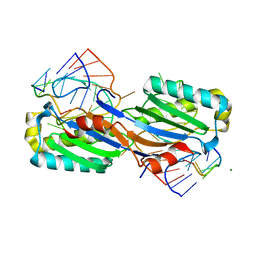 | | Crystal structure of the IS608 transposase in complex with left end 26-mer DNA hairpin and a 6-mer DNA representing the left end cleavage site | | Descriptor: | 5'-D(*DA*DA*DA*DG*DC*DC*DC*DC*DT*DA*DG*DC*DTP*DT *DT*DT*DA*DG*DC*DT*DA*DT*DG*DG*DG*DGP)-3', 5'-D(*DT*DA*DT*DT*DA*DCP)-3', MAGNESIUM ION, ... | | Authors: | Barabas, O, Ronning, D.R, Guynet, C, Hickman, A.B, Ton-Hoang, B, Chandler, M, Dyda, F. | | Deposit date: | 2007-12-13 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of is200/is605 Family DNA Transposases: Activation and Transposon-Directed Target Site Selection.
Cell(Cambridge,Mass.), 132, 2008
|
|
2VIC
 
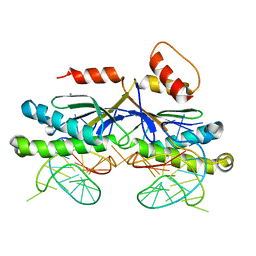 | | CRYSTAL STRUCTURE OF THE ISHP608 TRANSPOSASE IN COMPLEX with Left end 26- mer DNA and manganese | | Descriptor: | 5'-D(*AP*AP*AP*GP*CP*CP*CP*CP*TP*AP *GP*CP*TP*TP*TP*TP*AP*GP*CP*TP*AP*TP*GP*GP*GP*G)-3', MANGANESE (II) ION, TRANSPOSASE ORFA | | Authors: | Barabas, O, Ronning, D.R, Guynet, C, Hickman, A.B, Ton-Hoang, B, Chandler, M, Dyda, F. | | Deposit date: | 2007-11-29 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mechanism of is200/is605 Family DNA Transposases: Activation and Transposon-Directed Target Site Selection.
Cell(Cambridge,Mass.), 132, 2008
|
|
6W7G
 
 | | Structure of EED bound to inhibitor 1056 | | Descriptor: | 8-(2,6-dimethylpyridin-3-yl)-N-[(5-fluoro-2,3-dihydro-1-benzofuran-4-yl)methyl]-1-(methylsulfonyl)imidazo[1,5-c]pyrimidin-5-amine, FORMIC ACID, Polycomb protein EED, ... | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2020-03-19 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | EEDi-5285: An Exceptionally Potent, Efficacious, and Orally Active Small-Molecule Inhibitor of Embryonic Ectoderm Development.
J.Med.Chem., 63, 2020
|
|
4EUI
 
 | | Crystal Structure of MIF L46F mutant | | Descriptor: | Macrophage migration inhibitory factor, SULFATE ION | | Authors: | Ashrafi, A, Pojer, F, Lashuel, H. | | Deposit date: | 2012-04-25 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterization of molecular determinants of the conformational stability of macrophage migration inhibitory factor: leucine 46 hydrophobic pocket.
Plos One, 7, 2012
|
|
4XKR
 
 | | Crystal structure of NikA from Staphylococcus aureus in complex with Ni-(L-His)(2-methyl-thiazolidine dicarboxylate) (co-crystallization with Ni(II) and CDdeltaHis medium supernatant) | | Descriptor: | (2R,4R)-2-methyl-1,3-thiazolidine-2,4-dicarboxylic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Lebrette, H, Cavazza, C. | | Deposit date: | 2015-01-12 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Novel insights into nickel import in Staphylococcus aureus: the positive role of free histidine and structural characterization of a new thiazolidine-type nickel chelator.
Metallomics, 7, 2015
|
|
7SV2
 
 | | Human Cytochrome P450 (CYP) 3A5 ternary complex with azamulin | | Descriptor: | (3aS,4R,5S,6R,8R,9R,9aR,10R)-6-ethyl-5-hydroxy-4,6,9,10-tetramethyl-1-oxodecahydro-3a,9-propanocyclopenta[8]annulen-8-yl [(5-amino-1H-1,2,4-triazol-3-yl)sulfanyl]acetate, Cytochrome P450 3A5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hsu, M, Johnson, E.F. | | Deposit date: | 2021-11-18 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural characterization of the homotropic cooperative binding of azamulin to human cytochrome P450 3A5.
J.Biol.Chem., 298, 2022
|
|
4XKQ
 
 | |
2VIH
 
 | | CRYSTAL STRUCTURE OF THE IS608 TRANSPOSASE IN COMPLEX WITH Left END 26-MER DNA | | Descriptor: | 5'-D(*AP*AP*AP*GP*CP*CP*CP*CP*TP*AP *GP*CP*TP*TP*TP*TP*AP*GP*CP*TP*AP*TP*GP*GP*GP*G)-3', TRANSPOSASE ORFA | | Authors: | Barabas, O, Ronning, D.R, Guynet, C, Hickman, A.B, Ton-Hoang, B, Chandler, M, Dyda, F. | | Deposit date: | 2007-12-04 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of is200/is605 Family DNA Transposases: Activation and Transposon-Directed Target Site Selection.
Cell(Cambridge,Mass.), 132, 2008
|
|
5I95
 
 | | Crystal Structure of Human Mitochondrial Isocitrate Dehydrogenase R140Q Mutant Homodimer bound to NADPH and alpha-Ketoglutaric acid | | Descriptor: | 2-OXOGLUTARIC ACID, ACETATE ION, CALCIUM ION, ... | | Authors: | Zhang, B, Jin, L, Wu, W, Jiang, F, DeLaBarre, B, Travins, J.A, Padyana, A.K. | | Deposit date: | 2016-02-19 | | Release date: | 2017-03-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | AG-221, a First-in-Class Therapy Targeting Acute Myeloid Leukemia Harboring Oncogenic IDH2 Mutations.
Cancer Discov, 7, 2017
|
|
6WBZ
 
 | | Structure of Human HDAC2 in complex with an ethyl ketone inhibitor containing a spiro-bicyclic group | | Descriptor: | (1S)-N-{(1S)-7,7-dihydroxy-1-[5-(2-methoxyquinolin-3-yl)-1H-imidazol-2-yl]nonyl}-6-ethyl-6-azaspiro[2.5]octane-1-carboxamide, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Klein, D.J, Yu, W. | | Deposit date: | 2020-03-28 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Discovery of ethyl ketone-based HDACs 1, 2, and 3 selective inhibitors for HIV latency reactivation.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
5FPP
 
 | | Structure of a pre-reaction ternary complex between sarin- acetylcholinesterase and HI-6 | | Descriptor: | 2-(2-ETHOXYETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(AMINOCARBONYL)-1-[({2-[(E)-(HYDROXYIMINO)METHYL]PYRIDINIUM-1-YL}METHOXY)METHYL]PYRIDINIUM, ... | | Authors: | Allgardsson, A, Berg, L, Akfur, C, Hornberg, A, Worek, F, Linusson, A, Ekstrom, F. | | Deposit date: | 2015-12-02 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a Prereaction Complex between the Nerve Agent Sarin, its Biological Target Acetylcholinesterase, and the Antidote Hi-6.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8G66
 
 | | Structure with SJ3149 | | Descriptor: | (3S)-3-{5-[(1,2-benzoxazol-3-yl)amino]-1-oxo-1,3-dihydro-2H-isoindol-2-yl}piperidine-2,6-dione, Casein kinase I isoform alpha, DNA damage-binding protein 1, ... | | Authors: | Miller, D.J, Young, S.M, Fischer, M. | | Deposit date: | 2023-02-14 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Structure of ternary complex with molecular glue targeting CK1A for degradation by the CRL4CRBN ubiquitin ligase
To Be Published
|
|
5I96
 
 | | Crystal Structure of Human Mitochondrial Isocitrate Dehydrogenase (IDH2) R140Q Mutant Homodimer in Complex with AG-221 (Enasidenib) Inhibitor. | | Descriptor: | 2-methyl-1-[(4-[6-(trifluoromethyl)pyridin-2-yl]-6-{[2-(trifluoromethyl)pyridin-4-yl]amino}-1,3,5-triazin-2-yl)amino]propan-2-ol, ACETATE ION, CALCIUM ION, ... | | Authors: | Wei, W, Zhang, B, Jin, L, Jiang, F, DeLaBarre, B, Travins, J.A, Padyana, A.K. | | Deposit date: | 2016-02-19 | | Release date: | 2017-03-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | AG-221, a First-in-Class Therapy Targeting Acute Myeloid Leukemia Harboring Oncogenic IDH2 Mutations.
Cancer Discov, 7, 2017
|
|
8UTA
 
 | | yjdF riboswitch from R. gauvreauii in complex with proflavine bound to Fab BL3-6 S97N | | Descriptor: | Fab BL3-6 S97N heavy chain, Fab BL3-6 S97N light chain, MAGNESIUM ION, ... | | Authors: | Krochmal, D, Lewicka, A, Piccirilli, J.A. | | Deposit date: | 2023-10-30 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural basis for promiscuity in ligand recognition by yjdF riboswitch.
Cell Discov, 10, 2024
|
|
2VYE
 
 | | Crystal Structure of the DnaC-ssDNA complex | | Descriptor: | 5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP)-3', REPLICATIVE DNA HELICASE | | Authors: | Lo, Y.H, Tsai, K.L, Sun, Y.J, Hsiao, C.D. | | Deposit date: | 2008-07-23 | | Release date: | 2008-12-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | The Crystal Structure of a Replicative Hexameric Helicase Dnac and its Complex with Single-Stranded DNA.
Nucleic Acids Res., 37, 2009
|
|
