5VMO
 
 | | Crystal structure of grouper iridovirus GIV66:Bim complex | | Descriptor: | 1,2-ETHANEDIOL, Bak protein, Bcl-2 interacting mediator of cell death, ... | | Authors: | Banjara, S, Kvansakul, M. | | Deposit date: | 2017-04-28 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Grouper iridovirus GIV66 is a Bcl-2 protein that inhibits apoptosis by exclusively sequestering Bim.
J. Biol. Chem., 293, 2018
|
|
7CBK
 
 | | Structure of Human Neutrophil Elastase Ecotin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Ecotin, ... | | Authors: | Jobichen, C, Sivaraman, J. | | Deposit date: | 2020-06-12 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for the Inhibition Mechanism of Ecotin against Neutrophil Elastase by Targeting the Active Site and Secondary Binding Site.
Biochemistry, 59, 2020
|
|
3Q9L
 
 | | The structure of the dimeric E.coli MinD-ATP complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Septum site-determining protein minD | | Authors: | Wu, W, Park, K.-T, Lutkenhaus, J, Holyoak, T. | | Deposit date: | 2011-01-08 | | Release date: | 2011-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.343 Å) | | Cite: | Determination of the structure of the MinD-ATP complex reveals the orientation of MinD on the membrane and the relative location of the binding sites for MinE and MinC.
Mol.Microbiol., 79, 2011
|
|
5W3R
 
 | | SH2B1 SH2 Domain | | Descriptor: | PHENOL, PHOSPHATE ION, SH2B adapter protein 1 | | Authors: | McKercher, M.A, Wuttke, D.S. | | Deposit date: | 2017-06-08 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.386 Å) | | Cite: | Diversity in peptide recognition by the SH2 domain of SH2B1.
Proteins, 86, 2018
|
|
7C3F
 
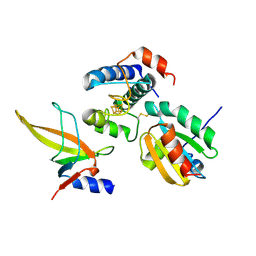 | | Crystal structure of ferredoxin: thioredoxin reductase and thioredoxin m2 complex | | Descriptor: | Ferredoxin-thioredoxin reductase catalytic chain, chloroplastic, Ferredoxin-thioredoxin reductase variable chain, ... | | Authors: | Kurisu, G, Juniar, L, Tanaka, H. | | Deposit date: | 2020-05-12 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3986 Å) | | Cite: | Structural basis for thioredoxin isoform-based fine-tuning of ferredoxin-thioredoxin reductase activity.
Protein Sci., 29, 2020
|
|
7CBP
 
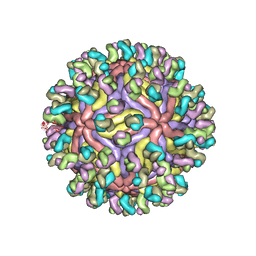 | | CryoEM structure of Zika virus with Fab at 4.1 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, Fab Heavy chain, ... | | Authors: | Tyagi, A, Ahmed, T, Shi, J, Bhushan, S. | | Deposit date: | 2020-06-13 | | Release date: | 2020-07-08 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | A complex between the Zika virion and the Fab of a broadly cross-reactive neutralizing monoclonal antibody revealed by cryo-EM and single particle analysis at 4.1 angstrom resolution.
J Struct Biol X, 4, 2020
|
|
7LHW
 
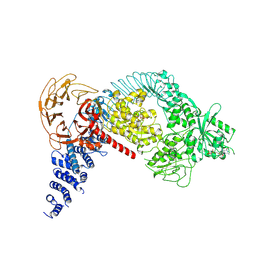 | | Structure of the LRRK2 monomer | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2 | | Authors: | Myasnikov, A, Zhu, H, Hixson, P, Xie, B, Yu, K, Pitre, A, Peng, J, Sun, J. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural analysis of the full-length human LRRK2.
Cell, 184, 2021
|
|
7LI3
 
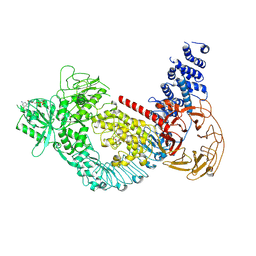 | | Structure of the LRRK2 G2019S mutant | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2 | | Authors: | Myasnikov, A, Zhu, H, Hixson, P, Xie, B, Yu, K, Pitre, A, Peng, J, Sun, J. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural analysis of the full-length human LRRK2.
Cell, 184, 2021
|
|
7LI4
 
 | | Structure of LRRK2 after symmetry expansion | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2 | | Authors: | Myasnikov, A, Zhu, H, Hixson, P, Xie, B, Yu, K, Pitre, A, Peng, J, Sun, J. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural analysis of the full-length human LRRK2.
Cell, 184, 2021
|
|
5W9A
 
 | | The structure of the Trim5alpha Bbox- coiled coil in complex LC3B | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B, Tripartite motif-containing protein 5, ZINC ION | | Authors: | Keown, J.R, Goldstone, D.C. | | Deposit date: | 2017-06-22 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | A helical LC3-interacting region mediates the interaction between the retroviral restriction factor Trim5 alpha and mammalian autophagy-related ATG8 proteins.
J. Biol. Chem., 293, 2018
|
|
3QMS
 
 | | Crystal structure of the mutant T159V,V182A,Y206F of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-02-05 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
7WUG
 
 | | GID subcomplex: Gid12 bound Substrate Receptor Scaffolding module | | Descriptor: | Glucose-induced degradation protein 8, HLJ1_G0042170.mRNA.1.CDS.1, Vacuolar import and degradation protein 24, ... | | Authors: | Qiao, S, Cheng, J.D, Schulman, B.A. | | Deposit date: | 2022-02-08 | | Release date: | 2022-06-08 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of Gid12-bound GID E3 reveal steric blockade as a mechanism inhibiting substrate ubiquitylation.
Nat Commun, 13, 2022
|
|
7LSU
 
 | | Ruminococcus bromii Amy12-D392A with 63-a-D-glucosyl-maltotriose | | Descriptor: | ACETATE ION, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Koropatkin, N.M, Cockburn, D.W, Brown, H.A, Kibler, R.D. | | Deposit date: | 2021-02-18 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and substrate recognition by the Ruminococcus bromii amylosome pullulanases.
J.Struct.Biol., 213, 2021
|
|
7LZ9
 
 | | Inactive form of VanR from S. coelicolor | | Descriptor: | MAGNESIUM ION, Putative two-component system response regulator | | Authors: | Maciunas, L.J, Loll, P.J. | | Deposit date: | 2021-03-09 | | Release date: | 2021-07-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of full-length VanR from Streptomyces coelicolor in both the inactive and activated states.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7LSA
 
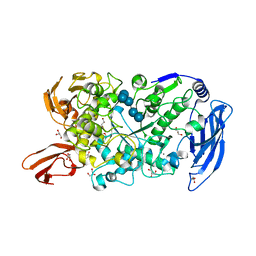 | | Ruminococcus bromii Amy12 with maltoheptaose | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Koropatkin, N.M, Cockburn, D.W, Brown, H.A, Kibler, R.D. | | Deposit date: | 2021-02-18 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure and substrate recognition by the Ruminococcus bromii amylosome pullulanases.
J.Struct.Biol., 213, 2021
|
|
7XKG
 
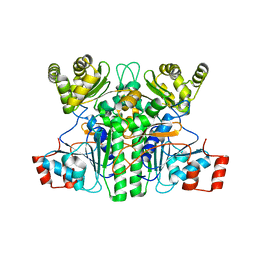 | | Crystal structure of an intramolecular mesacyl-CoA transferase from the 3-hydroxypropionic acid cycle of Roseiflexus castenholzii | | Descriptor: | Acyl-CoA transferase/carnitine dehydratase-like protein | | Authors: | Min, Z.Z, Fan, C.P, Wu, W.P, Xin, Y.Y, Liu, M.H, Zhang, X, Wang, Z.G, Xu, X.L. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of an Intramolecular Mesaconyl-Coenzyme A Transferase From the 3-Hydroxypropionic Acid Cycle of Roseiflexus castenholzii .
Front Microbiol, 13, 2022
|
|
7LST
 
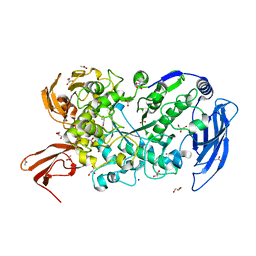 | | Ruminococcus bromii Amy12-D392A with 63-a-D-glucosyl-maltotriosyl-maltotriose | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Koropatkin, N.M, Cockburn, D.W, Brown, H.A, Kibler, R.D. | | Deposit date: | 2021-02-18 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and substrate recognition by the Ruminococcus bromii amylosome pullulanases.
J.Struct.Biol., 213, 2021
|
|
3QF0
 
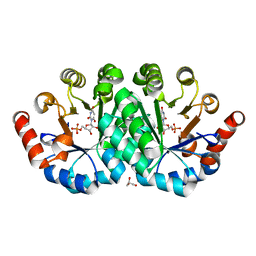 | | Crystal structure of the mutant T159V,Y206F of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-01-20 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
7LSR
 
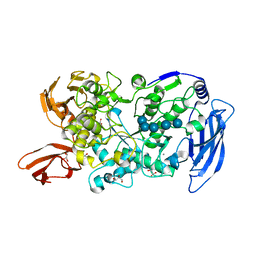 | | Ruminococcus bromii Amy12-D392A with maltoheptaose | | Descriptor: | CALCIUM ION, GLYCEROL, Pullulanase, ... | | Authors: | Koropatkin, N.M, Cockburn, D.W, Brown, H.A, Kibler, R.D. | | Deposit date: | 2021-02-18 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structure and substrate recognition by the Ruminococcus bromii amylosome pullulanases.
J.Struct.Biol., 213, 2021
|
|
7FJR
 
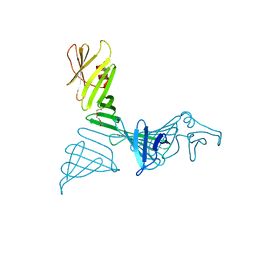 | | Structure of a mutant of OspA | | Descriptor: | DI(HYDROXYETHYL)ETHER, Outer surface protein A | | Authors: | Shiga, S, Makabe, K. | | Deposit date: | 2021-08-04 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | beta-Strand-mediated Domain-swapping in the Absence of Hydrophobic Core Repacking.
J.Mol.Biol., 436, 2024
|
|
7XN7
 
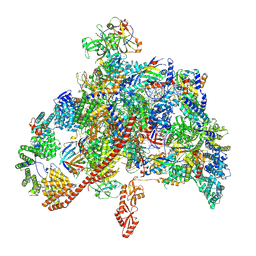 | | RNA polymerase II elongation complex containing Spt4/5, Elf1, Spt6, Spn1 and Paf1C | | Descriptor: | Chromatin elongation factor SPT5, Component of the Paf1p complex, Constituent of Paf1 complex with RNA polymerase II, ... | | Authors: | Ehara, H, Kujirai, T, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2022-04-28 | | Release date: | 2022-09-07 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of nucleosome disassembly and reassembly by RNAPII elongation complex with FACT.
Science, 377, 2022
|
|
7XB0
 
 | | Crystal structure of Omicron BA.2 RBD complexed with hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, L, Liao, H, Meng, Y, Li, W. | | Deposit date: | 2022-03-19 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of human ACE2 higher binding affinity to currently circulating Omicron SARS-CoV-2 sub-variants BA.2 and BA.1.1.
Cell, 185, 2022
|
|
4FX8
 
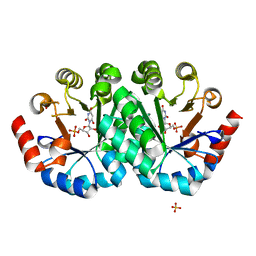 | | Crystal structure of the mutant Q185A.R203A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, Orotidine 5'-phosphate decarboxylase, SULFATE ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2012-07-02 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9411 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
7XAZ
 
 | | Crystal structure of Omicron BA.1.1 RBD complexed with hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Liao, H, Meng, Y, Li, W. | | Deposit date: | 2022-03-19 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of human ACE2 higher binding affinity to currently circulating Omicron SARS-CoV-2 sub-variants BA.2 and BA.1.1.
Cell, 185, 2022
|
|
7X39
 
 | | Structure of CIZ1 bound ERH | | Descriptor: | Enhancer of rudimentary homolog,Cip1-interacting zinc finger protein | | Authors: | Wang, X, Xu, C. | | Deposit date: | 2022-02-28 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular basis for the recognition of CIZ1 by ERH.
Febs J., 290, 2023
|
|
