1WOE
 
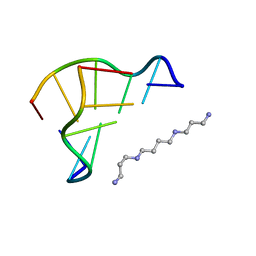 | | X-ray structure of a Z-DNA hexamer d(CGCGCG) | | Descriptor: | SPERMINE, Z-DNA hexamer | | Authors: | Chatake, T, Tanaka, I, Umino, H, Arai, S, Niimura, N. | | Deposit date: | 2004-08-15 | | Release date: | 2005-07-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The hydration structure of a Z-DNA hexameric duplex determined by a neutron diffraction technique.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1Q2N
 
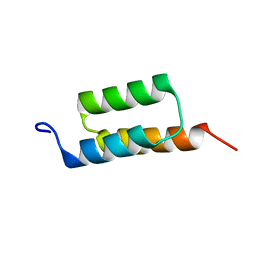 | |
7ELC
 
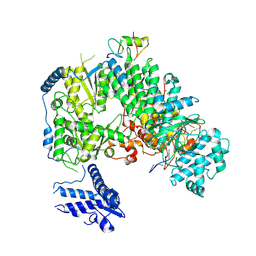 | | Structure of monomeric complex of MACV L bound to Z and 3'-vRNA | | Descriptor: | 3'-vRNA promoter, MANGANESE (II) ION, RING finger protein Z, ... | | Authors: | Xu, X, Peng, R, Peng, Q, Shi, Y. | | Deposit date: | 2021-04-09 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of Lassa and Machupo virus polymerases complexed with cognate regulatory Z proteins identify targets for antivirals.
Nat Microbiol, 6, 2021
|
|
1WW1
 
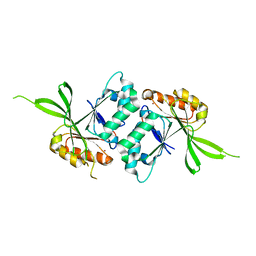 | |
1RC2
 
 | | 2.5 Angstrom Resolution X-ray Structure of Aquaporin Z | | Descriptor: | 2-O-octyl-beta-D-glucopyranose, Aquaporin Z | | Authors: | Savage, D.F, Egea, P.F, Robles, Y.C, O'Connell III, J.D, Stroud, R.M. | | Deposit date: | 2003-11-03 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Architecture and selectivity in aquaporins: 2.5 a X-ray structure of aquaporin Z
Plos Biol., 1, 2003
|
|
3H5C
 
 | | X-Ray Structure of Protein Z-Protein Z Inhibitor Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Protein Z-dependent protease inhibitor, Vitamin K-dependent protein Z, ... | | Authors: | Dementiev, A.A, Huang, X, Olson, S.T, Gettins, P.G.W. | | Deposit date: | 2009-04-21 | | Release date: | 2010-04-28 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | Basis for the specificity and activation of the serpin protein Z-dependent proteinase inhibitor (ZPI) as an inhibitor of membrane-associated factor Xa.
J.Biol.Chem., 285, 2010
|
|
1F66
 
 | | 2.6 A CRYSTAL STRUCTURE OF A NUCLEOSOME CORE PARTICLE CONTAINING THE VARIANT HISTONE H2A.Z | | Descriptor: | HISTONE H2A.Z, HISTONE H2B, HISTONE H3, ... | | Authors: | Suto, R.K, Clarkson, M.J, Tremethick, D.J, Luger, K. | | Deposit date: | 2000-06-20 | | Release date: | 2000-11-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a nucleosome core particle containing the variant histone H2A.Z.
Nat.Struct.Biol., 7, 2000
|
|
3EYI
 
 | |
2FK6
 
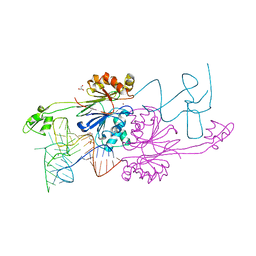 | | Crystal Structure of RNAse Z/tRNA(Thr) complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, RIBONUCLEASE Z, ... | | Authors: | Li de la Sierra-Gallay, I, Mathy, N, Pellegrini, O, Condon, C. | | Deposit date: | 2006-01-04 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the ubiquitous 3' processing enzyme RNase Z bound to transfer RNA.
Nat.Struct.Mol.Biol., 13, 2006
|
|
4M6B
 
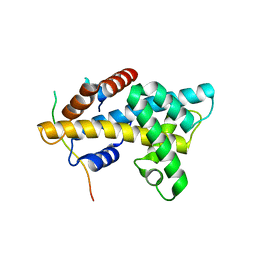 | | Crystal structure of yeast Swr1-Z domain in complex with H2A.Z-H2B dimer | | Descriptor: | Chimera protein of Histone H2B.1 and Histone H2A.Z, Helicase SWR1 | | Authors: | Hong, J.J, Feng, H.Q, Wang, F, Ranjan, A, Chen, J.H, Jiang, J.S, Girlando, R, Xiao, T.S, Wu, C, Bai, Y.W. | | Deposit date: | 2013-08-09 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The Catalytic Subunit of the SWR1 Remodeler Is a Histone Chaperone for the H2A.Z-H2B Dimer.
Mol.Cell, 53, 2014
|
|
4AFX
 
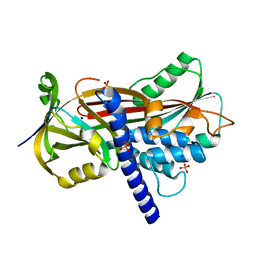 | | Crystal structure of the reactive loop cleaved ZPI in I2 space group | | Descriptor: | CALCIUM ION, PHOSPHATE ION, PROTEIN Z DEPENDENT PROTEASE INHIBITOR, ... | | Authors: | Zhou, A, Yan, Y, Wei, Z. | | Deposit date: | 2012-01-23 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural Basis for Catalytic Activation of Protein Z-Dependent Protease Inhibitor (Zpi) by Protein Z.
Blood, 120, 2012
|
|
4AJU
 
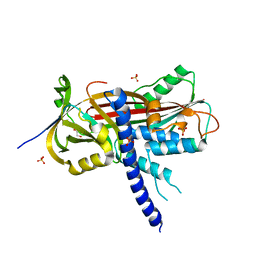 | | Crystal structure of the reactive loop cleaved ZPI in P41 space group | | Descriptor: | 1,2-ETHANEDIOL, PROTEIN Z-DEPENDENT PROTEASE INHIBITOR, SULFATE ION | | Authors: | Zhou, A, Yan, Y, Wei, Z. | | Deposit date: | 2012-02-19 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural Basis for Catalytic Activation of Protein Z-Dependent Protease Inhibitor (Zpi) by Protein Z.
Blood, 120, 2012
|
|
1Y44
 
 | | Crystal structure of RNase Z | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, PHOSPHATE ION, ... | | Authors: | de la Sierra-Gallay, I.L, Pellegrini, O, Condon, C. | | Deposit date: | 2004-11-30 | | Release date: | 2005-01-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for substrate binding, cleavage and allostery in the tRNA maturase RNase Z.
Nature, 433, 2005
|
|
2E7Y
 
 | | High resolution structure of T. maritima tRNase Z | | Descriptor: | S-1,2-PROPANEDIOL, SULFATE ION, ZINC ION, ... | | Authors: | Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-15 | | Release date: | 2007-09-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The structure of the flexible arm of Thermotoga maritima tRNase Z differs from those of homologous enzymes
Acta Crystallogr.,Sect.F, 63, 2007
|
|
4R15
 
 | | High-resolution crystal structure of Z-DNA in complex with Cr3+ cations | | Descriptor: | CHROMIUM ION, DNA (5'-D(*CP*GP*CP*GP*CP*G)-3') | | Authors: | Drozdzal, P, Gilski, M, Kierzek, R, Lomozik, L, Jaskolski, M. | | Deposit date: | 2014-08-04 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | High-resolution crystal structure of Z-DNA in complex with Cr(3+) cations.
J.Biol.Inorg.Chem., 20, 2015
|
|
3WA9
 
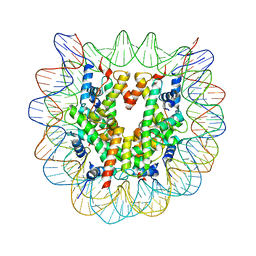 | | The nucleosome containing human H2A.Z.1 | | Descriptor: | DNA (146-MER), Histone H2A.Z, Histone H2B type 1-J, ... | | Authors: | Horikoshi, N, Sato, K, Shimada, K, Arimura, Y, Osakabe, A, Tachiwana, H, Iwasaki, W, Kagawa, W, Harata, M, Kimura, H, Kurumizaka, H. | | Deposit date: | 2013-04-30 | | Release date: | 2013-12-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Structural polymorphism in the L1 loop regions of human H2A.Z.1 and H2A.Z.2
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HIG
 
 | | Ultrahigh-resolution crystal structure of Z-DNA in complex with Mn2+ ion. | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*G)-3'), MANGANESE (II) ION, SPERMINE (FULLY PROTONATED FORM) | | Authors: | Drozdzal, P, Gilski, M, Kierzek, R, Lomozik, L, Jaskolski, M. | | Deposit date: | 2012-10-11 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.75 Å) | | Cite: | Ultrahigh-resolution crystal structures of Z-DNA in complex with Mn(2+) and Zn(2+) ions.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HIF
 
 | | Ultrahigh-resolution crystal structure of Z-DNA in complex with Zn2+ ions | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*GP*CP*GP*CP*G)-3'), SPERMINE (FULLY PROTONATED FORM), ... | | Authors: | Drozdzal, P, Gilski, M, Kierzek, R, Lomozik, L, Jaskolski, M. | | Deposit date: | 2012-10-11 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Ultrahigh-resolution crystal structures of Z-DNA in complex with Mn(2+) and Zn(2+) ions.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6OQU
 
 | | E. coli ATP synthase State 1d | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Stewart, A.G, Sobti, M, Walshe, J.L. | | Deposit date: | 2019-04-29 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures provide insight into how E. coli F1FoATP synthase accommodates symmetry mismatch.
Nat Commun, 11, 2020
|
|
6OQS
 
 | | E. coli ATP synthase State 1b | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Stewart, A.G, Sobti, M, Walshe, J.L. | | Deposit date: | 2019-04-29 | | Release date: | 2020-06-03 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures provide insight into how E. coli F1FoATP synthase accommodates symmetry mismatch.
Nat Commun, 11, 2020
|
|
7Z16
 
 | | E. coli C-P lyase bound to PhnK/PhnL dual ABC dimer with AMPPNP and PhnK E171Q mutation | | Descriptor: | Alpha-D-ribose 1-methylphosphonate 5-phosphate C-P lyase, Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnH, Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnI, ... | | Authors: | Amstrup, S.K, Sofus, N, Karlsen, J.L, Skjerning, R.B, Boesen, T, Enghild, J.J, Hove-Jensen, B, Brodersen, D.E. | | Deposit date: | 2022-02-24 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.09 Å) | | Cite: | Structural remodelling of the carbon-phosphorus lyase machinery by a dual ABC ATPase.
Nat Commun, 14, 2023
|
|
6WNQ
 
 | | E. coli ATP Synthase State 2a | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Stewart, A.G, Sobti, M, Walshe, J.L. | | Deposit date: | 2020-04-23 | | Release date: | 2020-06-03 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures provide insight into how E. coli F1FoATP synthase accommodates symmetry mismatch.
Nat Commun, 11, 2020
|
|
6WNR
 
 | | E. coli ATP synthase State 3b | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Stewart, A.G, Sobti, M, Walshe, J.L. | | Deposit date: | 2020-04-23 | | Release date: | 2020-06-03 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures provide insight into how E. coli F1FoATP synthase accommodates symmetry mismatch.
Nat Commun, 11, 2020
|
|
8IUM
 
 | | Cryo-EM structure of the tafluprost acid-bound human PTGFR-Gq complex | | Descriptor: | (~{Z})-7-[(1~{R},2~{R},3~{R},5~{S})-2-[(~{E})-3,3-bis(fluoranyl)-4-phenoxy-but-1-enyl]-3,5-bis(oxidanyl)cyclopentyl]hept-5-enoic acid, Antibody fragment scFv16, G subunit alpha (q), ... | | Authors: | Wu, C, Xu, Y, Xu, H.E. | | Deposit date: | 2023-03-24 | | Release date: | 2023-07-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Ligand-induced activation and G protein coupling of prostaglandin F 2 alpha receptor.
Nat Commun, 14, 2023
|
|
8IUK
 
 | | Cryo-EM structure of the PGF2-alpha-bound human PTGFR-Gq complex | | Descriptor: | (Z)-7-[(1R,2R,3R,5S)-3,5-bis(oxidanyl)-2-[(E,3S)-3-oxidanyloct-1-enyl]cyclopentyl]hept-5-enoic acid, Antibody fragment scFv16, G subunit alpha (q), ... | | Authors: | Wu, C, Xu, Y, Xu, H.E. | | Deposit date: | 2023-03-24 | | Release date: | 2023-07-12 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Ligand-induced activation and G protein coupling of prostaglandin F 2 alpha receptor.
Nat Commun, 14, 2023
|
|
