8SPX
 
 | | PS3 F1 Rotorless, high ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase subunit alpha, ... | | Authors: | Sobti, M, Stewart, A.G. | | Deposit date: | 2023-05-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | The series of conformational states adopted by rotorless F 1 -ATPase during its hydrolysis cycle.
Structure, 32, 2024
|
|
8HKG
 
 | |
8SWK
 
 | | Cryo-EM structure of NLRP3 closed hexamer | | Descriptor: | 1-[4-(2-oxidanylpropan-2-yl)furan-2-yl]sulfonyl-3-(1,2,3,5-tetrahydro-s-indacen-4-yl)urea, ADENOSINE-5'-TRIPHOSPHATE, NACHT, ... | | Authors: | Yu, X, Matico, R.E, Miller, R, Schoubroeck, B.V, Grauwen, K, Suarez, J, Pietrak, B, Haloi, N, Yin, Y, Tresadern, G.J, Perez-Benito, L, Lindahl, E, Bottelbergs, A, Oehlrich, D, Opdenbosch, N.V, Sharma, S. | | Deposit date: | 2023-05-18 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4.32 Å) | | Cite: | Cryo-EM structures of NLRP3 reveal its self-activation mechanism
Nat Commun, 2024
|
|
8H68
 
 | | Crystal structure of Caenorhabditis elegans NMAD-1 in complex with NOG and Mg(II) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shi, Y, Ding, J, Yang, H. | | Deposit date: | 2022-10-16 | | Release date: | 2023-02-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Caenorhabditis elegans NMAD-1 functions as a demethylase for actin.
J Mol Cell Biol, 15, 2023
|
|
8HHS
 
 | |
8HIL
 
 | | A cryo-EM structure of B. oleracea RNA polymerase V at 3.57 Angstrom | | Descriptor: | DNA-dependent RNA polymerase IV and V subunit 2, DNA-directed RNA polymerase V largest subunit, DNA-directed RNA polymerase subunit, ... | | Authors: | Du, X, Xie, G, Hu, H, Du, J. | | Deposit date: | 2022-11-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structure and mechanism of the plant RNA polymerase V.
Science, 379, 2023
|
|
8SXN
 
 | | Structure of NLRP3 and NEK7 complex | | Descriptor: | 1-[4-(2-oxidanylpropan-2-yl)furan-2-yl]sulfonyl-3-(1,2,3,5-tetrahydro-s-indacen-4-yl)urea, ADENOSINE-5'-TRIPHOSPHATE, NACHT, ... | | Authors: | Yu, X, Matico, R.E, Miller, R, Schoubroeck, B.V, Grauwen, K, Suarez, J, Pietrak, B, Haloi, N, Yin, Y, Tresadern, G.J, Perez-Benito, L, Lindahl, E, Bottelbergs, A, Oehlrich, D, Opdenbosch, N.V, Sharma, S. | | Deposit date: | 2023-05-22 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Cryo-EM structures of NLRP3 reveal its self-activation mechanism
Nat Commun, 2024
|
|
8SWF
 
 | | Cryo-EM structure of NLRP3 open octamer | | Descriptor: | NACHT, LRR and PYD domains-containing protein 3 | | Authors: | Yu, X, Matico, R.E, Miller, R, Schoubroeck, B.V, Grauwen, K, Suarez, J, Pietrak, B, Haloi, N, Yin, Y, Tresadern, G.J, Perez-Benito, L, Lindahl, E, Bottelbergs, A, Oehlrich, D, Opdenbosch, N.V, Sharma, S. | | Deposit date: | 2023-05-18 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Cryo-EM structures of NLRP3 reveal its self-activation mechanism
Nat Commun, 2024
|
|
8H3W
 
 | |
8SZH
 
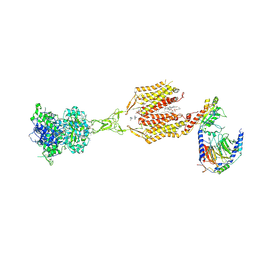 | | Cryo-EM structure of cinacalcet-bound human calcium-sensing receptor CaSR-Gi complex in lipid nanodiscs | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | He, F, Wu, C, Gao, Y, Skiniotis, G. | | Deposit date: | 2023-05-29 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Allosteric modulation and G-protein selectivity of the Ca 2+ -sensing receptor.
Nature, 626, 2024
|
|
8HLA
 
 | |
8HIM
 
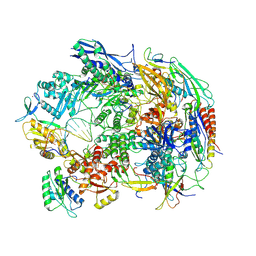 | | A cryo-EM structure of B. oleracea RNA polymerase V elongation complex at 2.73 Angstrom | | Descriptor: | DNA (34-MER), DNA-directed RNA polymerase IV and V subunit 2, DNA-directed RNA polymerase V largest subunit, ... | | Authors: | Hu, H, Xie, G, Du, X, Du, J. | | Deposit date: | 2022-11-21 | | Release date: | 2023-03-22 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure and mechanism of the plant RNA polymerase V.
Science, 379, 2023
|
|
8SGP
 
 | | human liver mitochondrial Medium-chain specific acyl-CoA dehydrogenase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Medium-chain specific acyl-CoA dehydrogenase, mitochondrial | | Authors: | Zhang, Z. | | Deposit date: | 2023-04-12 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | High-Resolution Structural Proteomics of Mitochondria Using the 'Build and Retrieve' Methodology.
Mol.Cell Proteomics, 22, 2023
|
|
8SGR
 
 | | human liver mitochondrial Isovaleryl-CoA dehydrogenase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Isovaleryl-CoA dehydrogenase, mitochondrial | | Authors: | Zhang, Z. | | Deposit date: | 2023-04-13 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | High-Resolution Structural Proteomics of Mitochondria Using the 'Build and Retrieve' Methodology.
Mol.Cell Proteomics, 22, 2023
|
|
8SGS
 
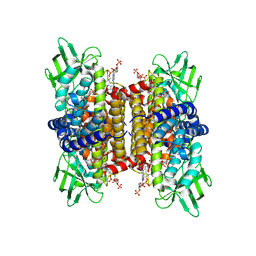 | | human liver mitochondrial Short-chain specific acyl-CoA dehydrogenase | | Descriptor: | COENZYME A, FLAVIN-ADENINE DINUCLEOTIDE, Short-chain specific acyl-CoA dehydrogenase, ... | | Authors: | Zhang, Z. | | Deposit date: | 2023-04-13 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | High-Resolution Structural Proteomics of Mitochondria Using the 'Build and Retrieve' Methodology.
Mol.Cell Proteomics, 22, 2023
|
|
8S32
 
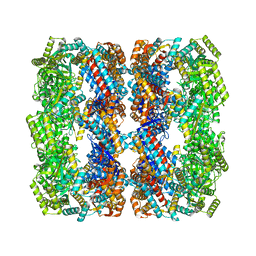 | | GroEL with bound GroTAC peptide | | Descriptor: | Chaperonin GroEL, GroTAC | | Authors: | Wroblewski, K, Izert-Nowakowska, M.A, Goral, T.K, Klimecka, M.M, Kmiecik, S, Gorna, M.W. | | Deposit date: | 2024-02-19 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Depletion of essential GroEL protein in Escherichia coli using Clp-Interacting Peptidic Protein Erasers (CLIPPERs)
To Be Published
|
|
8HCT
 
 | | Crystal structure of Cu2+ binding to Dendrorhynchus zhejiangensis ferritin | | Descriptor: | COPPER (II) ION, FE (III) ION, Ferritin, ... | | Authors: | Ming, T.H, Su, X.R, Huo, C.H. | | Deposit date: | 2022-11-03 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural and Biochemical Characterization of Silver/Copper Binding by Dendrorhynchus zhejiangensis Ferritin.
Polymers (Basel), 15, 2023
|
|
8SG4
 
 | |
8RZV
 
 | |
8H02
 
 | |
8SD8
 
 | | Carbonic anhydrase II radiation damage RT 91-120 | | Descriptor: | Carbonic anhydrase 2, ZINC ION | | Authors: | Combs, J.E, Mckenna, R. | | Deposit date: | 2023-04-06 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.789 Å) | | Cite: | XFEL structure of carbonic anhydrase II: a comparative study of XFEL, NMR, X-ray and neutron structures.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8SXF
 
 | |
8SNF
 
 | | Crystal structure of metformin hydrolase (MfmAB) from Pseudomonas mendocina sp. MET-2 with Ni2+2 bound | | Descriptor: | NICKEL (II) ION, metformin hydrolase subunit A, metformin hydrolase subunit B | | Authors: | Tassoulas, L.J, Rankin, J.A, Elias, M.H, Wackett, L.P. | | Deposit date: | 2023-04-27 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dinickel enzyme evolved to metabolize the pharmaceutical metformin and its implications for wastewater and human microbiomes.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SXG
 
 | |
8T0Z
 
 | | Human liver-type glutaminase (K253A) with L-Gln, filamentous form | | Descriptor: | GLUTAMINE, Glutaminase liver isoform, mitochondrial | | Authors: | Feng, S, Aplin, C, Nguyen, T.-T.T, Milano, S.K, Cerione, R.A. | | Deposit date: | 2023-06-01 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Filament formation drives catalysis by glutaminase enzymes important in cancer progression.
Nat Commun, 15, 2024
|
|
