3ONV
 
 | | Crystal structure of E. Coli purine nucleoside phosphorylase with SO4 | | Descriptor: | Purine nucleoside phosphorylase deoD-type, SULFATE ION | | Authors: | Mikleusevic, G, Stefanic, Z, Narzyk, M, Wielgus-Kutrowska, B, Bzowska, A, Luic, M. | | Deposit date: | 2010-08-30 | | Release date: | 2011-07-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | Validation of the catalytic mechanism of Escherichia coli purine nucleoside phosphorylase by structural and kinetic studies.
Biochimie, 93, 2011
|
|
1RS8
 
 | | Bovine endothelial NOS heme domain with D-lysine-D-nitroarginine amide bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, CACODYLATE ION, ... | | Authors: | Flinspach, M, Li, H, Jamal, J, Yang, W, Huang, H, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2003-12-09 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of the neuronal and endothelial nitric oxide synthase heme domain with D-nitroarginine-containing dipeptide inhibitors bound.
Biochemistry, 43, 2004
|
|
1RS6
 
 | | Rat neuronal NOS heme domain with D-lysine-D-nitroarginine amide bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, D-MANNITOL, ... | | Authors: | Flinspach, M, Li, H, Jamal, J, Yang, W, Huang, H, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2003-12-09 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the Neuronal and Endothelial Nitric Oxide Synthase Heme Domain with d-Nitroarginine-Containing Dipeptide Inhibitors Bound.
Biochemistry, 43, 2004
|
|
4R7A
 
 | | Crystal Structure of RBBP4 bound to PHF6 peptide | | Descriptor: | GLYCEROL, Histone-binding protein RBBP4, PHD finger protein 6 | | Authors: | Liu, Z, Li, F, Zhang, B, Li, S, Wu, J, Shi, Y. | | Deposit date: | 2014-08-27 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis of Plant Homeodomain Finger 6 (PHF6) Recognition by the Retinoblastoma Binding Protein 4 (RBBP4) Component of the Nucleosome Remodeling and Deacetylase (NuRD) Complex
J.Biol.Chem., 290, 2015
|
|
7EL9
 
 | | Structure of Machupo virus L polymerase in complex with Z protein and 3'-vRNA (dimeric complex) | | Descriptor: | MANGANESE (II) ION, Machupo virus 3'-vRNA promoter, RING finger protein Z, ... | | Authors: | Peng, R, Xu, X, Peng, Q, Shi, Y. | | Deposit date: | 2021-04-09 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of Lassa and Machupo virus polymerases complexed with cognate regulatory Z proteins identify targets for antivirals.
Nat Microbiol, 6, 2021
|
|
2O0F
 
 | | Docking of the modified RF3 X-ray structure into cryo-EM map of E.coli 70S ribosome bound with RF3 | | Descriptor: | Peptide chain release factor 3 | | Authors: | Gao, H, Zhou, Z, Rawat, U, Huang, C, Bouakaz, L, Wang, C, Liu, Y, Zavialov, A, Gursky, R, Sanyal, S, Ehrenberg, M, Frank, J, Song, H. | | Deposit date: | 2006-11-27 | | Release date: | 2007-07-24 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (15.5 Å) | | Cite: | RF3 induces ribosomal conformational changes responsible for dissociation of class I release factors
Cell(Cambridge,Mass.), 129, 2007
|
|
2G4F
 
 | | Structure of S.olivaceoviridis xylanase Q88A/R275A mutant | | Descriptor: | Hydrolase | | Authors: | Diertavitian, S, Kaneko, S, Fujimoto, Z, Kuno, A, Johansson, E, Lo Leggio, L. | | Deposit date: | 2006-02-22 | | Release date: | 2007-03-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure-based engineering of glucose specificity in a family 10 xylanase from Streptomyces olivaceoviridis E-86
PROCESS BIOCHEM, 47, 2012
|
|
3VOD
 
 | | Crystal Structure of mutant MarR C80S from E.coli | | Descriptor: | Multiple antibiotic resistance protein marR | | Authors: | Lou, H, Zhu, R, Hao, Z. | | Deposit date: | 2012-01-21 | | Release date: | 2013-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The multiple antibiotic resistance regulator MarR is a copper sensor in Escherichia coli.
Nat.Chem.Biol., 10, 2014
|
|
2G6G
 
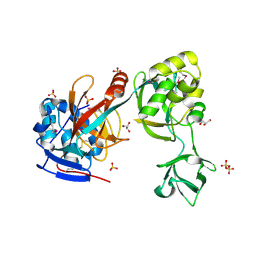 | | Crystal structure of MltA from Neisseria gonorrhoeae | | Descriptor: | GLYCEROL, GNA33, SULFATE ION | | Authors: | Powell, A.J, Liu, Z.J, Nicholas, R.A, Davies, C. | | Deposit date: | 2006-02-24 | | Release date: | 2006-05-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of the Lytic Transglycosylase MltA from N.gonorrhoeae and E.coli: Insights into Interdomain Movements and Substrate Binding.
J.Mol.Biol., 359, 2006
|
|
5BPE
 
 | | Crystal structure of EV71 3Cpro in complex with a potent and selective Inhibitor | | Descriptor: | (2~{S})-~{N}-[(1~{R},2~{S})-1-cyano-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepiperidin-3-yl]propan-2-yl]-3-phenyl-2-[[(~{E})-3-phenylprop-2-enoyl]amino]propanamide, EV71 3Cpro | | Authors: | Luqing, S, Yin, Z. | | Deposit date: | 2015-05-28 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cyanohydrin as an Anchoring Group for Potent and Selective Inhibitors of Enterovirus 71 3C Protease
J.Med.Chem., 58, 2015
|
|
3PX7
 
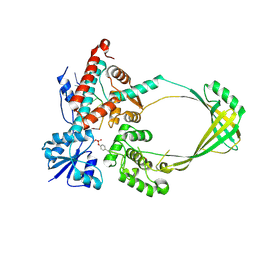 | |
4HXF
 
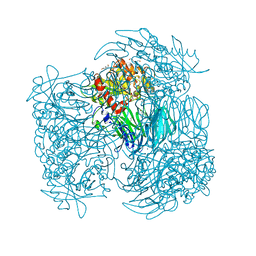 | | Acylaminoacyl peptidase in complex with Z-Gly-Gly-Phe-chloromethyl ketone | | Descriptor: | CHLORIDE ION, HEXANE-1,6-DIOL, MAGNESIUM ION, ... | | Authors: | Kiss-Szeman, A, Menyhard, D.K, Tichy-Racs, E, Hornung, B, Radi, K, Szeltner, Z, Domokos, K, Szamosi, I, Naray-Szabo, G, Polgar, L, Harmat, V. | | Deposit date: | 2012-11-09 | | Release date: | 2013-05-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | A Self-compartmentalizing Hexamer Serine Protease from Pyrococcus Horikoshii: SUBSTRATE SELECTION ACHIEVED THROUGH MULTIMERIZATION.
J.Biol.Chem., 288, 2013
|
|
2AMP
 
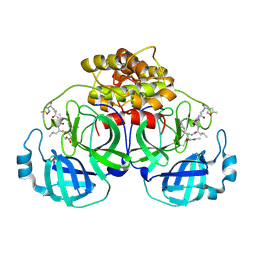 | | Crystal Structure Of Porcine Transmissible Gastroenteritis Virus Mpro in Complex with an Inhibitor N1 | | Descriptor: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]-L-ALANYL-L-VALYL-N~1~-((1S)-4-ETHOXY-4-OXO-1-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Yang, H, Xue, X, Yang, K, Zhao, Q, Bartlam, M, Rao, Z. | | Deposit date: | 2005-08-10 | | Release date: | 2005-09-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design of Wide-Spectrum Inhibitors Targeting Coronavirus Main Proteases.
Plos Biol., 3, 2005
|
|
4E04
 
 | | RpBphP2 chromophore-binding domain crystallized by homologue-directed mutagenesis. | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome (Light-regulated signal transduction histidine kinase), PhyB1 | | Authors: | Bellini, D, Papiz, M.Z. | | Deposit date: | 2012-03-02 | | Release date: | 2012-07-25 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Dimerization properties of the RpBphP2 chromophore-binding domain crystallized by homologue-directed mutagenesis.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5MTZ
 
 | | Crystal structure of a long form RNase Z from yeast | | Descriptor: | PHOSPHATE ION, Ribonuclease Z, ZINC ION | | Authors: | Li de la Sierra-Gallay, I, Miao, M, van Tilbeurgh, H. | | Deposit date: | 2017-01-11 | | Release date: | 2017-06-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The crystal structure of Trz1, the long form RNase Z from yeast.
Nucleic Acids Res., 45, 2017
|
|
8XMN
 
 | | Voltage-gated sodium channel Nav1.7 variant M2 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, N, Li, Z, Wu, Q, Huang, G. | | Deposit date: | 2023-12-27 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Dissection of the structure-function relationship of Na v channels.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8XMO
 
 | | Voltage-gated sodium channel Nav1.7 variant M4 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, N, Li, Z, Wu, Q, Huang, G. | | Deposit date: | 2023-12-27 | | Release date: | 2024-03-06 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Dissection of the structure-function relationship of Na v channels.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7CUT
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with Z-VAD-FMK | | Descriptor: | 3C protein, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Lu, M, Yang, H.T, Wang, Z.Y, Zhao, Y, Xing, Y.F. | | Deposit date: | 2020-08-24 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Identification of proteasome and caspase inhibitors targeting SARS-CoV-2 M pro .
Signal Transduct Target Ther, 6, 2021
|
|
7DRT
 
 | | Human Wntless in complex with Wnt3a | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhong, Q, Zhao, Y, Ye, F, Xiao, Z, Huang, G, Zhang, Y, Lu, P, Xu, W, Zhou, Q, Ma, D. | | Deposit date: | 2020-12-29 | | Release date: | 2021-07-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Cryo-EM structure of human Wntless in complex with Wnt3a.
Nat Commun, 12, 2021
|
|
3QNW
 
 | | Caspase-6 in complex with Z-VAD-FMK inhibitor | | Descriptor: | Caspase-6, Z-VAD-FMK | | Authors: | Mueller, I, Lamers, M, Ritchie, A, Park, H, Dominguez, C, Munoz, I, Maillard, M, Kiselyov, A. | | Deposit date: | 2011-02-09 | | Release date: | 2011-09-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of human caspase-6 in complex with Z-VAD-FMK: New peptide binding mode observed for the non-canonical caspase conformation.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
7BQY
 
 | | THE CRYSTAL STRUCTURE OF COVID-19 MAIN PROTEASE IN COMPLEX WITH AN INHIBITOR N3 at 1.7 angstrom | | Descriptor: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Liu, X, Zhang, B, Jin, Z, Yang, H, Rao, Z. | | Deposit date: | 2020-03-26 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Mprofrom SARS-CoV-2 and discovery of its inhibitors.
Nature, 582, 2020
|
|
2AOA
 
 | | Crystal structures of a high-affinity macrocyclic peptide mimetic in complex with the Grb2 SH2 domain | | Descriptor: | 2-(4-((9S,10S,14S,Z)-18-(2-AMINO-2-OXOETHYL)-9-(CARBOXYMETHYL)-14-(NAPHTHALEN-1-YLMETHYL)-8,17,20-TRIOXO-7,16,19-TRIAZASPIRO[5.14]ICOS-11-EN-10-YL)PHENYL)MALONIC ACID, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Growth factor receptor-bound protein 2 | | Authors: | Phan, J, Shi, Z.D, Burke, T.R, Waugh, D.S. | | Deposit date: | 2005-08-12 | | Release date: | 2005-10-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structures of a High-affinity Macrocyclic Peptide Mimetic in Complex with the Grb2 SH2 Domain.
J.Mol.Biol., 353, 2005
|
|
4JBA
 
 | |
4M6B
 
 | | Crystal structure of yeast Swr1-Z domain in complex with H2A.Z-H2B dimer | | Descriptor: | Chimera protein of Histone H2B.1 and Histone H2A.Z, Helicase SWR1 | | Authors: | Hong, J.J, Feng, H.Q, Wang, F, Ranjan, A, Chen, J.H, Jiang, J.S, Girlando, R, Xiao, T.S, Wu, C, Bai, Y.W. | | Deposit date: | 2013-08-09 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The Catalytic Subunit of the SWR1 Remodeler Is a Histone Chaperone for the H2A.Z-H2B Dimer.
Mol.Cell, 53, 2014
|
|
4CAY
 
 | | Crystal structure of a human Anp32e-H2A.Z-H2B complex | | Descriptor: | ACIDIC LEUCINE-RICH NUCLEAR PHOSPHOPROTEIN 32 FAMILY MEMBER E, HISTONE H2A.Z, HISTONE H2B TYPE 1-J | | Authors: | Obri, A, Ouararhni, K, Papin, C, Diebold, M.-L, Padmanabhan, K, Marek, M, Stoll, I, Roy, L, Reilly, P.T, Mak, T.W, Dimitrov, S, Romier, C, Hamiche, A. | | Deposit date: | 2013-10-09 | | Release date: | 2014-01-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Anp32E is a Histone Chaperone that Removes H2A.Z from Chromatin
Nature, 648, 2014
|
|
