5A1P
 
 | |
2J4W
 
 | | Structure of a Plasmodium vivax apical membrane antigen 1-Fab F8.12.19 complex | | Descriptor: | APICAL MEMBRANE ANTIGEN 1, FAB FRAGMENT OF MONOCLONAL ANTIBODY F8.12.19 | | Authors: | Igonet, S, Vulliez-Le Normand, B, Faure, G, Riottot, M.M, Kocken, C.H.M, Thomas, A.W, Bentley, G.A. | | Deposit date: | 2006-09-07 | | Release date: | 2007-01-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cross-Reactivity Studies of an Anti-Plasmodium Vivax Apical Membrane Antigen 1 Monoclonal Antibody: Binding and Structural Characterisation.
J.Mol.Biol., 366, 2007
|
|
6JV4
 
 | | Crystal structure of metallo-beta-lactamase VMB-1 | | Descriptor: | CITRIC ACID, VMB-1, ZINC ION | | Authors: | Cheng, Q, Chen, S. | | Deposit date: | 2019-04-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Genetic and Biochemical Characterization of VMB-1, a Novel Metallo-beta-Lactamase Encoded by a Conjugative, Broad-Host Range IncC Plasmid from Vibrio spp.
Adv Biosyst, 4, 2020
|
|
5RW0
 
 | | PanDDA analysis group deposition -- Crystal Structure of DHTKD1 in complex with Z2444997446 | | Descriptor: | 1-{(2S,4S)-4-fluoro-1-[(2-methyl-1,3-thiazol-4-yl)methyl]pyrrolidin-2-yl}methanamine, MAGNESIUM ION, Probable 2-oxoglutarate dehydrogenase E1 component DHKTD1, ... | | Authors: | Bezerra, G.A, Foster, W.R, Bailey, H.J, Shrestha, L, Krojer, T, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Yue, W.W. | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.673 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7BVP
 
 | | AdhE spirosome in extended conformation | | Descriptor: | Aldehyde-alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Kim, G.J, Song, J.J. | | Deposit date: | 2020-04-11 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Aldehyde-alcohol dehydrogenase undergoes structural transition to form extended spirosomes for substrate channeling.
Commun Biol, 3, 2020
|
|
5AKQ
 
 | | X-ray structure and mutagenesis studies of the N-isopropylammelide isopropylaminohydrolase, AtzC | | Descriptor: | CHLORIDE ION, N-ISOPROPYLAMMELIDE ISOPROPYL AMIDOHYDROLASE, ZINC ION | | Authors: | Balotra, S, Warden, A.C, Newman, J, Briggs, L.J, Scott, C, Peat, T.S. | | Deposit date: | 2015-03-05 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-Ray Structure and Mutagenesis Studies of the N-Isopropylammelide Isopropylaminohydrolase, Atzc
Plos One, 1, 2015
|
|
6DA2
 
 | | Human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2018-05-01 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure-Activity Relationships of Rationally Designed Ritonavir Analogues: Impact of Side-Group Stereochemistry, Headgroup Spacing, and Backbone Composition on the Interaction with CYP3A4.
Biochemistry, 58, 2019
|
|
6DA5
 
 | | Human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, tert-butyl [(2R)-1-{[(2S)-1-oxo-3-phenyl-1-{[(pyridin-3-yl)methyl]amino}propan-2-yl]sulfanyl}-3-phenylpropan-2-yl]carbamate | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2018-05-01 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Activity Relationships of Rationally Designed Ritonavir Analogues: Impact of Side-Group Stereochemistry, Headgroup Spacing, and Backbone Composition on the Interaction with CYP3A4.
Biochemistry, 58, 2019
|
|
6DAG
 
 | | Human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, tert-butyl [(2R)-1-{[(2S)-1-oxo-3-phenyl-1-{[2-(pyridin-3-yl)ethyl]amino}propan-2-yl]sulfanyl}-3-phenylpropan-2-yl]carbamate | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2018-05-01 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Activity Relationships of Rationally Designed Ritonavir Analogues: Impact of Side-Group Stereochemistry, Headgroup Spacing, and Backbone Composition on the Interaction with CYP3A4.
Biochemistry, 58, 2019
|
|
5RGB
 
 | | Crystal Structure of Kemp Eliminase HG3.3b with bound transition state analogue, 277K | | Descriptor: | 6-NITROBENZOTRIAZOLE, Kemp Eliminase HG3.3b, SULFATE ION | | Authors: | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | Deposit date: | 2020-03-19 | | Release date: | 2020-07-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
3VMS
 
 | | Crystal structure of Staphylococcus aureus membrane-bound transglycosylase in complex with NBD-Lipid II | | Descriptor: | Monofunctional glycosyltransferase | | Authors: | Huang, C.Y, Shih, H.W, Lin, L.Y, Tien, Y.W, Cheng, T.J.R, Cheng, W.C, Wong, C.H, Ma, C. | | Deposit date: | 2011-12-15 | | Release date: | 2012-04-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Crystal structure of Staphylococcus aureus transglycosylase in complex with a lipid II analog and elucidation of peptidoglycan synthesis mechanism
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8X6M
 
 | | Crystal Structure of Glycerol Dehydrogenase in the Presence of NAD+ and Glycerol | | Descriptor: | GLYCEROL, Glycerol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Park, T, Kang, J.Y, Jin, M, Yang, J, Kim, H, Noh, C, Eom, S.H. | | Deposit date: | 2023-11-21 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the octamerization of glycerol dehydrogenase.
Plos One, 19, 2024
|
|
6YCS
 
 | | Human Transcription Cofactor PC4 DNA-binding domain in complex with full phosphorothioate 5-10-5 2'-O-methyl DNA gapmer antisense oligonucleotide. | | Descriptor: | DNA (5'-D(P*(OKQ))-D(P*(OKT))-R(P*(RFJ))-D(*(OKQ)P*(OKT)P*(AS)P*(GS)P*(OKN)P*(OKN)P*(PST)P*(OKN)P*(PST)P*(GS)P*(GS)P*(AS)P*(OKT)P*(OKT))-3'), PC4 protein, SODIUM ION, ... | | Authors: | Hyjek-Skladanowska, M, Vickers, T.A, Napiorkowska, A, Anderson, B, Tanowitz, M, Crooke, S.T, Liang, X, Seth, P.P, Nowotny, M. | | Deposit date: | 2020-03-19 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Origins of the Increased Affinity of Phosphorothioate-Modified Therapeutic Nucleic Acids for Proteins.
J.Am.Chem.Soc., 142, 2020
|
|
5A5J
 
 | | Cytochrome 2C9 P450 inhibitor complex | | Descriptor: | CYTOCHROME P450 2C9, N-[4-(3-chloranyl-4-cyano-phenoxy)-3,5-dimethoxy-phenyl]-1,1,1-tris(fluoranyl)methanesulfonamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Skerratt, S.E, de Groot, M.J, Phillips, C. | | Deposit date: | 2015-06-18 | | Release date: | 2016-08-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of a Novel Binding Pocket for Cyp 2C9 Inhibitors: Crystallography, Pharmacophore Modelling and Inhibitor Sar.
To be Published
|
|
2M55
 
 | | NMR structure of the complex of an N-terminally acetylated alpha-synuclein peptide with calmodulin | | Descriptor: | Alpha-synuclein, CALCIUM ION, Calmodulin | | Authors: | Gruschus, J.M, Yap, T, Pistolesi, S, Maltsev, A.S, Lee, J.C. | | Deposit date: | 2013-02-13 | | Release date: | 2013-05-08 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Calmodulin Complexed to an N-Terminally Acetylated alpha-Synuclein Peptide.
Biochemistry, 52, 2013
|
|
6DVV
 
 | | 2.25 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Klebsiella pneumoniae in Complex with NAD and Mn2+. | | Descriptor: | 6-phospho-alpha-glucosidase, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Endres, M, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-06-25 | | Release date: | 2018-07-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
6YC8
 
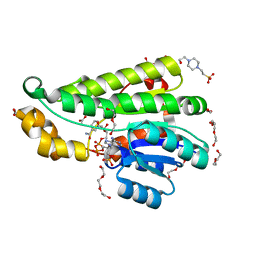 | | Crystal structure of KRED1-Pglu enzyme | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Di Pisa, F. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural insights into the desymmetrization of bulky 1,2-dicarbonyls through enzymatic monoreduction.
Bioorg.Chem., 108, 2021
|
|
2HRK
 
 | |
5RGD
 
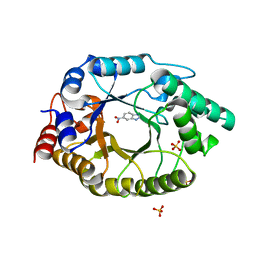 | | Crystal Structure of Kemp Eliminase HG3.14 with bound transition state analogue, 277K | | Descriptor: | 6-NITROBENZOTRIAZOLE, Kemp Eliminase HG3.14, SULFATE ION | | Authors: | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | Deposit date: | 2020-03-19 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
2J5L
 
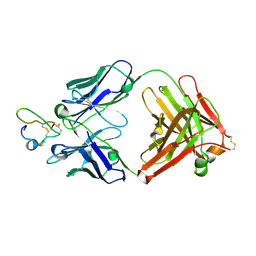 | | Structure of a Plasmodium falciparum apical membrane antigen 1-Fab F8. 12.19 complex | | Descriptor: | APICAL MEMBRANE ANTIGEN 1, FAB FRAGMENT OF MONOCLONAL ANTIBODY F8.12.19 | | Authors: | Igonet, S, Vulliez-Le Normand, B, Faure, G, Riottot, M.M, Kocken, C.H.M, Thomas, A.W, Bentley, G.A. | | Deposit date: | 2006-09-18 | | Release date: | 2007-01-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cross-Reactivity Studies of an Anti-Plasmodium Vivax Apical Membrane Antigen 1 Monoclonal Antibody: Binding and Structural Characterisation.
J.Mol.Biol., 366, 2007
|
|
5RG4
 
 | | Crystal Structure of Kemp Eliminase HG3 in unbound state, 277K | | Descriptor: | ACETATE ION, Kemp Eliminase HG3, SULFATE ION | | Authors: | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | Deposit date: | 2020-03-19 | | Release date: | 2020-07-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
6DXP
 
 | | The crystal structure of an FMN-dependent NADH-azoreductase from Klebsiella pneumoniae | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase | | Authors: | Arcinas, A.J, Ghosh, A, Chamala, S, Bonanno, J.B, Kelly, L, Almo, S.C. | | Deposit date: | 2018-06-29 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.478 Å) | | Cite: | The crystal structure of an FMN-dependent NADH-azoreductase from Klebsiella pneumoniae
To Be Published
|
|
5RW1
 
 | | PanDDA analysis group deposition of ground-state model of DHTKD1 | | Descriptor: | MAGNESIUM ION, Probable 2-oxoglutarate dehydrogenase E1 component DHKTD1, mitochondrial, ... | | Authors: | Bezerra, G.A, Foster, W.R, Bailey, H.J, Shrestha, L, Krojer, T, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Yue, W.W. | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5RGC
 
 | | Crystal Structure of Kemp Eliminase HG3.7 with bound transition state analogue, 277K | | Descriptor: | 6-NITROBENZOTRIAZOLE, Kemp Eliminase HG3, SULFATE ION | | Authors: | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | Deposit date: | 2020-03-19 | | Release date: | 2020-07-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
2DR6
 
 | | Crystal structure of a multidrug transporter reveal a functionally rotating mechanism | | Descriptor: | ACRB, DOXORUBICIN | | Authors: | Murakami, S, Nakashima, R, Yamashita, E, Matsumoto, T. | | Deposit date: | 2006-06-08 | | Release date: | 2006-08-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structures of a multidrug transporter reveal a functionally rotating mechanism
Nature, 443, 2006
|
|
