3PXT
 
 | | Crystal Structure of Ferrous CO Adduct of MauG in Complex with Pre-Methylamine Dehydrogenase | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, ACETATE ION, CALCIUM ION, ... | | Authors: | Yukl, E.T, Goblirsch, B.R, Wilmot, C.M. | | Deposit date: | 2010-12-10 | | Release date: | 2011-03-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal Structures of CO and NO Adducts of MauG in Complex with Pre-Methylamine Dehydrogenase: Implications for the Mechanism of Dioxygen Activation.
Biochemistry, 50, 2011
|
|
4MZ8
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with an Internal Deletion of CBS Domain from Campylobacter jejuni complexed with inhibitor compound C91 | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Gorla, S.K, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-29 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5004 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Campylobacter jejuni complexed with inhibitor compound C91
To be Published
|
|
3H3X
 
 | | Structure of the V74M large subunit mutant of NI-FE hydrogenase in an oxidized state | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Volbeda, A, Martinez, N, Martin, L, Fontecilla-Camps, J.C. | | Deposit date: | 2009-04-17 | | Release date: | 2009-07-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Introduction of methionines in the gas channel makes [NiFe] hydrogenase aero-tolerant
J.Am.Chem.Soc., 131, 2009
|
|
1HXN
 
 | |
7V2E
 
 | | Active state complex I from Q10-NADH dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-08-08 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5OS3
 
 | | Crystal structure of Aurora-A kinase in complex with an allosterically binding fragment | | Descriptor: | (1~{R})-1-(4-ethoxyphenyl)ethanamine, ADENOSINE-5'-DIPHOSPHATE, Aurora kinase A, ... | | Authors: | McIntyre, P.J, Collins, P.M, von Delft, F, Bayliss, R. | | Deposit date: | 2017-08-16 | | Release date: | 2017-11-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Characterization of Three Druggable Hot-Spots in the Aurora-A/TPX2 Interaction Using Biochemical, Biophysical, and Fragment-Based Approaches.
ACS Chem. Biol., 12, 2017
|
|
7F8O
 
 | | Cryo-EM structure of the C-terminal deletion mutant of human PANX1 in a nanodisc | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Pannexin-1 | | Authors: | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2021-07-02 | | Release date: | 2022-01-26 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
4I9H
 
 | | Crystal structure of rabbit LDHA in complex with AP28669 | | Descriptor: | 1-O-[3-(5-carboxypyridin-2-yl)-5-fluorophenyl]-6-O-[4-({[(5-carboxypyridin-2-yl)sulfanyl]acetyl}amino)-2-chloro-5-methoxyphenyl]-D-mannitol, L-lactate dehydrogenase A chain | | Authors: | Zhou, T, Stephan, Z.G, Kohlmann, A, Li, F, Commodore, L, Greenfield, M.T, Shakespeare, W.C, Zhu, X, Dalgarno, D.C. | | Deposit date: | 2012-12-05 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Fragment growing and linking lead to novel nanomolar lactate dehydrogenase inhibitors.
J.Med.Chem., 56, 2013
|
|
2KEW
 
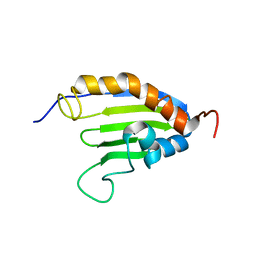 | | The solution structure of Bacillus subtilis SR211 START domain by NMR spectroscopy | | Descriptor: | Uncharacterized protein yndB | | Authors: | Mercier, K.A, Mueller, G.A, Powers, R, Acton, T.B, Ciano, M, Ho, C, Lui, J, Ma, L, Rost, B, Rossi, R, Xiao, R, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-02-05 | | Release date: | 2009-03-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | (1)H, (13)C, and (15)N NMR assignments for the Bacillus subtilis yndB START domain.
Biomol.Nmr Assign., 3, 2009
|
|
6M73
 
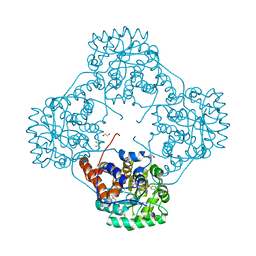 | | Crystal structure of Enterococcus hirae L-lactate oxidase in complex with D-lactate form ligand | | Descriptor: | 1,2-ETHANEDIOL, 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Yoshida, H, Hiraka, K, Tsugawa, W, Sode, K. | | Deposit date: | 2020-03-16 | | Release date: | 2021-03-17 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of lactate oxidase from Enterococcus hirae revealed new aspects of active site loop function: Product-inhibition mechanism and oxygen gatekeeper
Protein Sci., 31, 2022
|
|
6M74
 
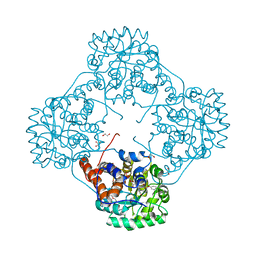 | | Crystal structure of Enterococcus hirae L-lactate oxidase M207L in complex with D-lactate form ligand | | Descriptor: | 1,2-ETHANEDIOL, 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Yoshida, H, Hiraka, K, Tsugawa, W, Sode, K. | | Deposit date: | 2020-03-16 | | Release date: | 2021-03-17 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structure of lactate oxidase from Enterococcus hirae revealed new aspects of active site loop function: Product-inhibition mechanism and oxygen gatekeeper
Protein Sci., 31, 2022
|
|
4QOP
 
 | | Structure of Bacillus pumilus catalase with hydroquinone bound. | | Descriptor: | CHLORIDE ION, Catalase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2014-06-20 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unprecedented access of phenolic substrates to the heme active site of a catalase: Substrate binding and peroxidase-like reactivity of Bacillus pumilus catalase monitored by X-ray crystallography and EPR spectroscopy.
Proteins, 83, 2015
|
|
4QJ1
 
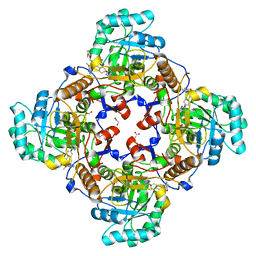 | | Co-crystal structure of the catalytic domain of the inosine monophosphate dehydrogenase from Cryptosporidium parvum with inhibitor N109 | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, FORMIC ACID, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-06-03 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.415 Å) | | Cite: | Co-crystal structure of the catalytic domain of the inosine monophosphate dehydrogenase from Cryptosporidium parvum with inhibitor N109
To be Published
|
|
4BB3
 
 | | Isopenicillin N synthase with the dipeptide substrate analogue AhC | | Descriptor: | (2S)-2-azanyl-6-oxidanylidene-6-[[(2S)-1-oxidanyl-1-oxidanylidene-4-sulfanyl-butan-2-yl]amino]hexanoic acid, FE (III) ION, ISOPENICILLIN N SYNTHASE, ... | | Authors: | Daruzzaman, A, Clifton, I.J, Rutledge, P.J. | | Deposit date: | 2012-09-19 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Crystal Structure of Isopenicillin N Synthase with a Dipeptide Substrate Analogue.
Arch.Biochem.Biophys., 530, 2013
|
|
3HBB
 
 | | Structures of dihydrofolate reductase-thymidylate synthase of Trypanosoma cruzi in the folate-free state and in complex with two antifolate drugs, trimetrexate and methotrexate | | Descriptor: | 1,2-ETHANEDIOL, Dihydrofolate reductase-thymidylate synthase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schormann, N, Senkovich, O, Chattopadhyay, D. | | Deposit date: | 2009-05-04 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of dihydrofolate reductase-thymidylate synthase of Trypanosoma cruzi in the folate-free state and in complex with two antifolate drugs, trimetrexate and methotrexate.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
4QI0
 
 | | X-ray structure of the ROQ domain from murine Roquin-1 | | Descriptor: | 1,2-ETHANEDIOL, Roquin-1 | | Authors: | Janowski, R, Schlundt, A, Sattler, M, Niessing, D. | | Deposit date: | 2014-05-30 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural basis for RNA recognition in roquin-mediated post-transcriptional gene regulation.
Nat.Struct.Mol.Biol., 21, 2014
|
|
5DKB
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a 3-methylphenylamino-substituted ethyl triaryl-ethylene derivative 4,4'-(2-{3-[(3-methylphenyl)amino]phenyl}but-1-ene-1,1-diyl)diphenol | | Descriptor: | 4,4'-(2-{3-[(3-methylphenyl)amino]phenyl}but-1-ene-1,1-diyl)diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-03 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
6LZN
 
 | | Thermolysin | | Descriptor: | CALCIUM ION, GLYCEROL, ISOLEUCINE, ... | | Authors: | Nam, K.H. | | Deposit date: | 2020-02-19 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural analysis of metal chelation of the metalloproteinase thermolysin by 1,10-phenanthroline.
J.Inorg.Biochem., 215, 2021
|
|
4FAV
 
 | |
5EJ8
 
 | | EcMenD-ThDP-Mn2+ complex structure soaked with 2-ketoglutarate for 2 min | | Descriptor: | (4S)-4-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3lambda~5~-thiazol-2-yl}-4-hydroxybutanoic acid, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Song, H.G, Dong, C, Chen, Y.Z, Sun, Y.R, Guo, Z.H. | | Deposit date: | 2015-11-01 | | Release date: | 2016-06-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | A Thiamine-Dependent Enzyme Utilizes an Active Tetrahedral Intermediate in Vitamin K Biosynthesis
J.Am.Chem.Soc., 138, 2016
|
|
2W2E
 
 | | 1.15 Angstrom crystal structure of P.pastoris aquaporin, Aqy1, in a closed conformation at pH 3.5 | | Descriptor: | AQUAPORIN PIP2-7 7, CHLORIDE ION, octyl beta-D-glucopyranoside | | Authors: | Fischer, G, Kosinska-Eriksson, U, Aponte-Santamaria, C, Palmgren, M, Geijer, C, Hedfalk, K, Hohmann, S, de Groot, B.L, Neutze, R, Lindkvist-Petersson, K. | | Deposit date: | 2008-10-29 | | Release date: | 2009-06-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal Structure of a Yeast Aquaporin at 1.15 A Reveals a Novel Gating Mechanism.1.15 A
Plos Biol., 7, 2009
|
|
3IG6
 
 | | Low molecular weigth human Urokinase type Plasminogen activator 2-[6-(3'-Aminomethyl-biphenyl-3-yloxy)-4-(3-dimethylamino-pyrrolidin-1-yl)-3,5-difluoro-pyridin-2-yloxy]-4-dimethylamino-benzoic acid complex | | Descriptor: | 2-[(6-{[3'-(aminomethyl)biphenyl-3-yl]oxy}-4-[(3R)-3-(dimethylamino)pyrrolidin-1-yl]-3,5-difluoropyridin-2-yl)oxy]-4-(dimethylamino)benzoic acid, PHOSPHATE ION, Urokinase-type plasminogen activator | | Authors: | Adler, M, Whitlow, M. | | Deposit date: | 2009-07-27 | | Release date: | 2009-10-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Identification of orally bioavailable, non-amidine inhibitors of Urokinase Plasminogen Activator (uPA)
Bioorg.Med.Chem.Lett., 19, 2009
|
|
1UXY
 
 | | MURB MUTANT WITH SER 229 REPLACED BY ALA, COMPLEX WITH ENOLPYRUVYL-UDP-N-ACETYLGLUCOSAMINE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, URIDINE DIPHOSPHO-N-ACETYLENOLPYRUVYLGLUCOSAMINE REDUCTASE, URIDINE-DIPHOSPHATE-2(N-ACETYLGLUCOSAMINYL) BUTYRIC ACID | | Authors: | Benson, T.E, Walsh, C.T, Hogle, J.M. | | Deposit date: | 1996-11-08 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structures of the S229A mutant and wild-type MurB in the presence of the substrate enolpyruvyl-UDP-N-acetylglucosamine at 1.8-A resolution.
Biochemistry, 36, 1997
|
|
7UKT
 
 | | Integrin alpha IIB beta3 complex with BMS4.2 | | Descriptor: | (1-{[(5S)-3-(4-carbamimidoylphenyl)-4,5-dihydro-1,2-oxazol-5-yl]methyl}piperidin-4-yl)acetic acid, 10E5 Fab heavy chain, 10E5 Fab light chain, ... | | Authors: | Lin, F.-Y, Zhu, J, Zhu, J, Springer, T.A. | | Deposit date: | 2022-04-01 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.36994433 Å) | | Cite: | A general chemical principle for creating closure-stabilizing integrin inhibitors.
Cell, 185, 2022
|
|
1VBI
 
 | |
