5XHN
 
 | | Crystal structure of Frog M-ferritin K104E mutant | | Descriptor: | CHLORIDE ION, Ferritin, middle subunit, ... | | Authors: | Jagdev, M.K, Vasudevan, D. | | Deposit date: | 2017-04-21 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Surface charge dependent separation of modified and hybrid ferritin in native PAGE: Impact of lysine 104
Biochim. Biophys. Acta, 1865, 2017
|
|
7UZD
 
 | |
7UDQ
 
 | | Crystal structure of COQ8A-CA157 inhibitor complex in space group P1 | | Descriptor: | 4-[(3,4,5-trimethoxyphenyl)amino]quinoline-7-carbonitrile, Atypical kinase COQ8A, mitochondrial, ... | | Authors: | Bingman, C.A, Murray, N, Smith, R.W, Pagliarini, D.J. | | Deposit date: | 2022-03-20 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Small-molecule inhibition of the archetypal UbiB protein COQ8.
Nat.Chem.Biol., 19, 2023
|
|
5XHW
 
 | | Crystal structure of HddC from Yersinia pseudotuberculosis | | Descriptor: | Putative 6-deoxy-D-mannoheptose pathway protein, SULFATE ION | | Authors: | Park, J, Kim, H, Kim, S, Shin, D.H. | | Deposit date: | 2017-04-24 | | Release date: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of d-glycero-alpha-d-manno-heptose-1-phosphate guanylyltransferase from Yersinia pseudotuberculosis.
Biochim. Biophys. Acta, 1866, 2018
|
|
6N2A
 
 | |
7UU6
 
 | |
7UU8
 
 | |
7UUC
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI19 | | Descriptor: | 3C-like proteinase nsp5, N-[(benzyloxy)carbonyl]-3-methyl-L-valyl-3-cyclopropyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | | Authors: | Yang, K.S, Liu, W.R. | | Deposit date: | 2022-04-28 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Novel Y-Shaped, S-O-N-O-S-Bridged Cross-Link between Three Residues C22, C44, and K61 Is Frequently Observed in the SARS-CoV-2 Main Protease.
Acs Chem.Biol., 18, 2023
|
|
7UU7
 
 | |
7UUB
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI12 | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, N-[(benzyloxy)carbonyl]-L-valyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methylidene-L-norvalinamide | | Authors: | Yang, K.S, Liu, W.R. | | Deposit date: | 2022-04-28 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | A Novel Y-Shaped, S-O-N-O-S-Bridged Cross-Link between Three Residues C22, C44, and K61 Is Frequently Observed in the SARS-CoV-2 Main Protease.
Acs Chem.Biol., 18, 2023
|
|
5FH6
 
 | | Crystal structure of the fifth bromodomain of human PB1 in complex with compound 10 | | Descriptor: | (3~{R})-3-(piperidin-1-ylmethyl)-2,3-dihydro-1~{H}-pyrrolo[1,2-a]quinazolin-5-one, Protein polybromo-1 | | Authors: | Tallant, C, Sutherell, C.L, Siejka, P, Krojer, T, Picaud, S, Fonseca, M, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Ley, S.V, Knapp, S. | | Deposit date: | 2015-12-21 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification and Development of 2,3-Dihydropyrrolo[1,2-a]quinazolin-5(1H)-one Inhibitors Targeting Bromodomains within the Switch/Sucrose Nonfermenting Complex.
J.Med.Chem., 59, 2016
|
|
5JBD
 
 | | 4,6-alpha-glucanotransferase GTFB from Lactobacillus reuteri 121 | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Pijning, T, Dijkstra, B.W, Bai, Y, Gangoiti-Munecas, J, Dijkhuizen, L. | | Deposit date: | 2016-04-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of 4,6-alpha-Glucanotransferase Supports Diet-Driven Evolution of GH70 Enzymes from alpha-Amylases in Oral Bacteria.
Structure, 25, 2017
|
|
5FHF
 
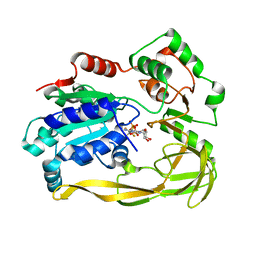 | | Crystal structure of Bacteroides sp Pif1 in complex with ADP-AlF4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, TETRAFLUOROALUMINATE ION, Uncharacterized protein | | Authors: | Zhou, X, Ren, W, Bharath, S.R, Song, H. | | Deposit date: | 2015-12-22 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Functional Insights into the Unwinding Mechanism of Bacteroides sp Pif1
Cell Rep, 14, 2016
|
|
5XAL
 
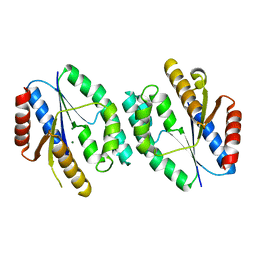 | |
7UUA
 
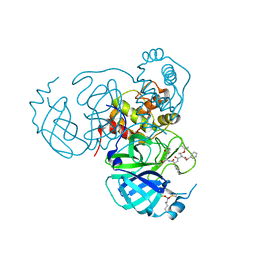 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI8 | | Descriptor: | 3C-like proteinase nsp5, N-[(BENZYLOXY)CARBONYL]-O-(TERT-BUTYL)-L-THREONYL-3-CYCLOHEXYL-N-[(1S)-2-HYDROXY-1-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}ETHYL]-L-ALANINAMIDE | | Authors: | Yang, K.S, Liu, W.R. | | Deposit date: | 2022-04-28 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Novel Y-Shaped, S-O-N-O-S-Bridged Cross-Link between Three Residues C22, C44, and K61 Is Frequently Observed in the SARS-CoV-2 Main Protease.
Acs Chem.Biol., 18, 2023
|
|
5J5B
 
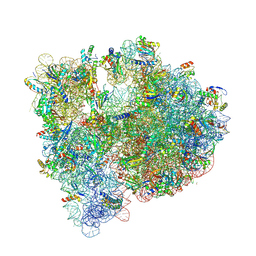 | | Structure of the WT E coli ribosome bound to tetracycline | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1,4-DIAMINOBUTANE, ... | | Authors: | Cocozaki, A, Ferguson, A. | | Deposit date: | 2016-04-01 | | Release date: | 2016-07-27 | | Last modified: | 2018-08-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Resistance mutations generate divergent antibiotic susceptibility profiles against translation inhibitors.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6MGE
 
 | | Structure of human 4-1BBL | | Descriptor: | GLYCEROL, PHOSPHATE ION, Tumor necrosis factor ligand superfamily member 9 | | Authors: | Kimberlin, C.R, Chin, S.M, Roe-Zurz, Z, Xu, A, Yang, Y. | | Deposit date: | 2018-09-13 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of the 4-1BB/4-1BBL complex and distinct binding and functional properties of utomilumab and urelumab.
Nat Commun, 9, 2018
|
|
7UU9
 
 | |
5FAJ
 
 | |
8Q5Z
 
 | |
6MH9
 
 | | The crystal structure of the Staphylococcus aureus Fatty acid Kinase (Fak) B1 protein A121I mutant to 2.02 Angstrom resolution | | Descriptor: | Fatty Acid Kinase (Fak) B1 protein, PALMITIC ACID | | Authors: | Cuypers, M.G, Ericson, M, Subramanian, C, White, S.W, Rock, C.O. | | Deposit date: | 2018-09-17 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Identification of structural transitions in bacterial fatty acid binding proteins that permit ligand entry and exit at membranes.
J.Biol.Chem., 298, 2022
|
|
7V9E
 
 | | Crystal structure of a methyl transferase ribozyme | | Descriptor: | BARIUM ION, GUANINE, RNA (68-MER), ... | | Authors: | Deng, J, Lilley, D.M.J, Huang, L. | | Deposit date: | 2021-08-25 | | Release date: | 2022-03-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanism of a methyltransferase ribozyme.
Nat.Chem.Biol., 18, 2022
|
|
5JEE
 
 | | Apo-structure of humanised RadA-mutant humRadA26F | | Descriptor: | CALCIUM ION, DNA repair and recombination protein RadA | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-18 | | Release date: | 2016-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
7VL4
 
 | | The complex structure of beta-1,2-glucosyltransferase from Ignavibacterium album with methyl beta-D-glucoside | | Descriptor: | CALCIUM ION, beta-1,2-glucosyltransferase, methyl beta-D-glucopyranoside | | Authors: | Kobayashi, K, Shimizu, H, Tanaka, N, Kuramochi, K, Nakai, H, Nakajima, M, Taguchi, H. | | Deposit date: | 2021-10-01 | | Release date: | 2022-03-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Characterization and structural analyses of a novel glycosyltransferase acting on the beta-1,2-glucosidic linkages.
J.Biol.Chem., 298, 2022
|
|
5XE1
 
 | | Crystal structure of the indoleamine 2,3-dioxygenagse 1 (IDO1) complexed with INCB14943 | | Descriptor: | 4-Amino-N-(3-chloro-4-fluorophenyl)-N'-hydroxy-1,2,5-oxadiazole-3-carboxamidine, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Xu, J, Wu, U, Liu, J. | | Deposit date: | 2017-03-31 | | Release date: | 2017-05-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into the binding mechanism of IDO1 with hydroxylamidine based inhibitor INCB14943
Biochem. Biophys. Res. Commun., 487, 2017
|
|
