3SBE
 
 | | Crystal structure of RAC1 P29S mutant | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Ha, B.H, Boggon, T.J. | | Deposit date: | 2011-06-03 | | Release date: | 2012-07-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma.
Nat.Genet., 44, 2012
|
|
3S45
 
 | | wild-type HIV-2 protease with antiviral drug amprenavir | | Descriptor: | CHLORIDE ION, IMIDAZOLE, Protease, ... | | Authors: | Tie, Y.-F, Wang, Y.-F, Weber, I.T. | | Deposit date: | 2011-05-18 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Critical differences in HIV-1 and HIV-2 protease specificity for clinical inhibitors.
Protein Sci., 21, 2012
|
|
3S9K
 
 | | Crystal structure of the Itk SH2 domain. | | Descriptor: | CITRIC ACID, Tyrosine-protein kinase ITK/TSK | | Authors: | Joseph, R.E, Ginder, N.D, Hoy, J.A, Nix, J.C, Fulton, B.D, Honzatko, R.B, Andreotti, A.H. | | Deposit date: | 2011-06-01 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.354 Å) | | Cite: | Structure of the interleukin-2 tyrosine kinase Src homology 2 domain; comparison between X-ray and NMR-derived structures.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3SFC
 
 | | Structure-Based Optimization of Potent 4- and 6-Azaindole-3-Carboxamides as Renin Inhibitors | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Renin, ... | | Authors: | Scheiper, B, Matter, H, Steinhagen, H, Bocskei, Z, Fleury, V, McCort, G. | | Deposit date: | 2011-06-13 | | Release date: | 2011-08-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based optimization of potent 4- and 6-azaindole-3-carboxamides as renin inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3SBK
 
 | | Russell's viper venom serine proteinase, RVV-V (PPACK-bound form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, Vipera russelli proteinase RVV-V gamma | | Authors: | Nakayama, D, Ben Ammar, Y, Takeda, S. | | Deposit date: | 2011-06-05 | | Release date: | 2011-09-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis of coagulation factor V recognition for cleavage by RVV-V
Febs Lett., 585, 2011
|
|
7WL3
 
 | | CVB5 expended empty particle | | Descriptor: | Capsid protein, Genome polyprotein | | Authors: | Yang, P, Wang, K. | | Deposit date: | 2022-01-12 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Atomic Structures of Coxsackievirus B5 Provide Key Information on Viral Evolution and Survival.
J.Virol., 96, 2022
|
|
7WM3
 
 | | hnRNP A2/B1 RRMs in complex with single-stranded DNA | | Descriptor: | DNA (5'-D(P*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*G)-3'), Heterogeneous nuclear ribonucleoproteins A2/B1 | | Authors: | Abula, A, Liu, Y, Guo, H, Li, T, Ji, X. | | Deposit date: | 2022-01-14 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural Insight Into hnRNP A2/B1 Homodimerization and DNA Recognition.
J.Mol.Biol., 435, 2022
|
|
7WQL
 
 | | Bovin Beta-lactoglobulin binding with zinc ions | | Descriptor: | Beta-lactoglobulin, ZINC ION | | Authors: | Li, T, Ma, J, Zang, J, Zhao, G, Zhang, T. | | Deposit date: | 2022-01-25 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Zinc binding strength of proteins dominants zinc uptake in Caco-2 cells.
Rsc Adv, 12, 2022
|
|
7WQG
 
 | | Bovin Alpha-lactalbumin binding with zinc ions | | Descriptor: | Alpha-lactalbumin, ZINC ION | | Authors: | Li, T, Zhang, T. | | Deposit date: | 2022-01-25 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Zinc binding strength of proteins dominants zinc uptake in Caco-2 cells.
Rsc Adv, 12, 2022
|
|
7WEX
 
 | |
7WHU
 
 | | Human Neutrophil Elastase in-complex with Ecotin Peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Ecotin Peptide, ... | | Authors: | Shankar, S, Jayaraman, S. | | Deposit date: | 2021-12-31 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Sequence preference and scaffolding requirement for the inhibition of human neutrophil elastase by ecotin peptide
Protein Sci., 31, 2022
|
|
3R1A
 
 | | Closed crystal structure of cytochrome P450 2B4 covalently bound to the mechanism-based inactivator tert-butylphenylacetylene | | Descriptor: | (4-tert-butylphenyl)acetaldehyde, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gay, S.C, Zhang, H, Stout, C.D, Hollenberg, P.F, Halpert, J.R. | | Deposit date: | 2011-03-09 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural Analysis of Mammalian Cytochrome P450 2B4 Covalently Bound to the Mechanism-Based Inactivator tert-Butylphenylacetylene: Insight into Partial Enzymatic Activity.
Biochemistry, 50, 2011
|
|
3R8Q
 
 | |
3R9B
 
 | | Crystal structure of Mycobacterium smegmatis CYP164A2 in ligand free state | | Descriptor: | 1,2-ETHANEDIOL, CYTOCHROME P450 164A2, DODECANE, ... | | Authors: | Agnew, C.R.J, Warrilow, A.G.S, Kelly, S.L, Brady, R.L. | | Deposit date: | 2011-03-25 | | Release date: | 2012-04-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | An enlarged, adaptable active site in CYP164 family P450 enzymes, the sole P450 in Mycobacterium leprae.
Antimicrob.Agents Chemother., 56, 2012
|
|
7W46
 
 | | Crystal structure of Bacillus subtilis YjoB with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Uncharacterized ATPase YjoB | | Authors: | Dahal, P, Kwon, E, Kim, D.Y. | | Deposit date: | 2021-11-26 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and biochemical analysis suggest that YjoB ATPase is a putative substrate-specific molecular chaperone.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3RRY
 
 | | H-Ras crosslinked control, soaked in aqueous solution: one of 10 in MSCS set | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Mattos, C, Buhrman, G, Kearney, B. | | Deposit date: | 2011-05-02 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Analysis of Binding Site Hot Spots on the Surface of Ras GTPase.
J.Mol.Biol., 413, 2011
|
|
3RS1
 
 | | Mouse C-type lectin-related protein Clrg | | Descriptor: | C-type lectin domain family 2 member I, CHLORIDE ION | | Authors: | Skalova, T, Dohnalek, J, Duskova, J, Stepankova, A, Koval, T, Hasek, J, Kotynkova, K, Vanek, O, Bezouska, K. | | Deposit date: | 2011-05-02 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mouse Clr-g, a Ligand for NK Cell Activation Receptor NKR-P1F: Crystal Structure and Biophysical Properties.
J.Immunol., 189, 2012
|
|
3RSO
 
 | | H-Ras soaked in 20% S,R,S-bisfuranol: 1 of 10 in MSCS set | | Descriptor: | (3S,3aR,6aS)-hexahydrofuro[2,3-b]furan-3-ol, CALCIUM ION, GTPase HRas, ... | | Authors: | Mattos, C, Buhrman, G, Kearney, B. | | Deposit date: | 2011-05-02 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Analysis of Binding Site Hot Spots on the Surface of Ras GTPase.
J.Mol.Biol., 413, 2011
|
|
3RS4
 
 | | H-Ras soaked in 60% 1,6-hexanediol: 1 of 10 in MSCS set | | Descriptor: | CALCIUM ION, GTPase HRas, HEXANE-1,6-DIOL, ... | | Authors: | Mattos, C, Buhrman, G, Kearney, B. | | Deposit date: | 2011-05-02 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Analysis of Binding Site Hot Spots on the Surface of Ras GTPase.
J.Mol.Biol., 413, 2011
|
|
7W42
 
 | | Crystal structure of Bacillus subtilis YjoB | | Descriptor: | Uncharacterized ATPase YjoB | | Authors: | Dahal, P, Kwon, E, Kim, D.Y. | | Deposit date: | 2021-11-26 | | Release date: | 2022-10-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.619 Å) | | Cite: | Crystal structure and biochemical analysis suggest that YjoB ATPase is a putative substrate-specific molecular chaperone.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5O5M
 
 | | Crystal Structure of the Protein-Kinase A catalytic subunit from Criteculus Griseus in complex with compounds RKp120 and RKp117 | | Descriptor: | CHLORIDE ION, RKp120, [2-[(4-isoquinolin-5-ylsulfonyl-1,4-diazepan-1-yl)methyl]phenyl]boronic acid, ... | | Authors: | Mueller, J.M, Heine, A, Klebe, G. | | Deposit date: | 2017-06-02 | | Release date: | 2018-06-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.583 Å) | | Cite: | Conceptional Design of Self-Assembling Bisubstrate-like Inhibitors of Protein Kinase A Resulting in a Boronic Acid Glutamate Linkage
Acs Omega, 2019
|
|
3RAB
 
 | | GPPNHP-BOUND RAB3A AT 2.0 A RESOLUTION | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, PROTEIN (RAB3A) | | Authors: | Dumas, J.J, Zhu, Z, Connolly, J.L, Lambright, D.G. | | Deposit date: | 1998-11-16 | | Release date: | 1999-04-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of activation and GTP hydrolysis in Rab proteins.
Structure Fold.Des., 7, 1999
|
|
7WCQ
 
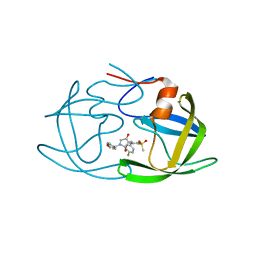 | | Crystal structure of HIV-1 protease in complex with lactam derivative 1 | | Descriptor: | (3R,4R)-3-[(4-fluorophenyl)methyl]-1-[(4-methoxyphenyl)methyl]-3-(4-methylsulfonylphenyl)-4-oxidanyl-pyrrolidin-2-one, Protease | | Authors: | Kojima, E, Iimuro, A, Nakajima, M, Kinuta, H, Asada, N, Sako, Y, Nakata, Z, Uemura, K, Arita, S, Miki, S, Wakasa-Morimoto, C, Tachibana, Y, Fumoto, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Pocket-to-Lead: Structure-Based De Novo Design of Novel Non-peptidic HIV-1 Protease Inhibitors Using the Ligand Binding Pocket as a Template.
J.Med.Chem., 65, 2022
|
|
5ODT
 
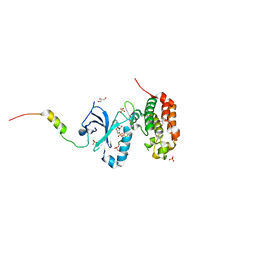 | | Aurora-A in complex with TACC3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Aurora kinase A, GLYCEROL, ... | | Authors: | Burgess, S.G, Bayliss, R. | | Deposit date: | 2017-07-06 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.021 Å) | | Cite: | Mitotic spindle association of TACC3 requires Aurora-A-dependent stabilization of a cryptic alpha-helix.
EMBO J., 37, 2018
|
|
7WBS
 
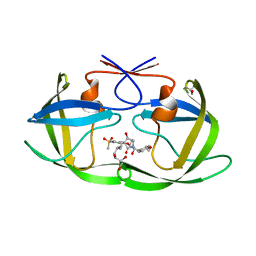 | | Crystal structure of HIV-1 protease in complex with lactam derivative 2 | | Descriptor: | (3~{R},4~{R})-1-[(4-methoxyphenyl)methyl]-3-(3-methylbutyl)-3-[4-methylsulfonyl-2-[(2~{S})-1-oxidanylpropan-2-yl]oxy-phenyl]-4-oxidanyl-pyrrolidin-2-one, GLYCEROL, Protease | | Authors: | Kojima, E, Iimuro, A, Nakajima, M, Kinuta, H, Asada, N, Sako, Y, Nakata, Z, Uemura, K, Arita, S, Miki, S, Wakabayashi-Morimoto, C, Tachibana, Y, Fumoto, M. | | Deposit date: | 2021-12-17 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Pocket-to-Lead: Structure-Based De Novo Design of Novel Non-peptidic HIV-1 Protease Inhibitors Using the Ligand Binding Pocket as a Template.
J.Med.Chem., 65, 2022
|
|
