5IN7
 
 | | X-RAY STRUCTURE OF THE N-TERMINAL DOMAIN OF HUMAN DOUBLECORTIN | | Descriptor: | Neuronal migration protein doublecortin | | Authors: | Ruf, A, Benz, J, Burger, D, D'Arcy, B, Debulpaep, M, Di Lello, P, Fry, D, Huber, W, Kremer, T, Laeremans, T, Matile, H, Ross, A, Rudolph, M.G, Rufer, A.C, Sharma, A, Steinmetz, M.O, Steyaert, J, Schoch, G, Stihle, M, Thoma, R. | | Deposit date: | 2016-03-07 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal Structures of the Human Doublecortin C- and N-terminal Domains in Complex with Specific Antibodies.
J.Biol.Chem., 291, 2016
|
|
4JFK
 
 | | Increasing the Efficiency Efficiency of Ligands for the FK506-Binding Protein 51 by Conformational Control: Complex of FKBP51 with (1S,6R)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-10-[(2-oxo-2,3-dihydro-1,3-benzothiazol-6-yl)sulfonyl]-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,6R)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-10-[(2-oxo-2,3-dihydro-1,3-benzothiazol-6-yl)sulfonyl]-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Wang, Y, Kirschner, A, Fabian, A, Gopalakrishnan, R, Kress, C, Hoogeland, B, Koch, U, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2013-02-28 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Increasing the efficiency of ligands for FK506-binding protein 51 by conformational control.
J.Med.Chem., 56, 2013
|
|
5IEP
 
 | | Structure of CDL2.3b, a computationally designed Vitamin-D3 binder | | Descriptor: | 3-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANOL, CDL2.3b | | Authors: | Stoddard, B.L, Doyle, L.A. | | Deposit date: | 2016-02-25 | | Release date: | 2017-03-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | Unintended specificity of an engineered ligand-binding protein facilitated by unpredicted plasticity of the protein fold.
Protein Eng.Des.Sel., 31, 2018
|
|
7EZT
 
 | | The structure and functional mechanism of nucleotide regulated acetylhexosaminidase Am2136 from Akkermansia muciniphila | | Descriptor: | Beta-N-acetylhexosaminidase, MAGNESIUM ION | | Authors: | Bao, R, Li, C.C, Tang, X.Y, Zhu, Y.B, Song, Y.J, Zhao, N.L, Huang, Q, Mou, X.Y, Luo, G.H, Liu, T.G. | | Deposit date: | 2021-06-02 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Nucleotide binding as an allosteric regulatory mechanism for Akkermansia muciniphila beta- N -acetylhexosaminidase Am2136.
Gut Microbes, 14, 2022
|
|
2OPC
 
 | | Structure of Melampsora lini avirulence protein, AvrL567-A | | Descriptor: | AvrL567-A, COBALT (II) ION, IMIDAZOLE | | Authors: | Guncar, G, Wang, C.I, Forwood, J.K, Teh, T, Catanzariti, A.M, Ellis, J.G, Dodds, P.N, Kobe, B. | | Deposit date: | 2007-01-29 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The use of Co2+ for crystallization and structure determination, using a conventional monochromatic X-ray source, of flax rust avirulence protein.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
3FCI
 
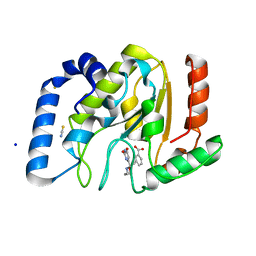 | | Complex of UNG2 and a fragment-based designed inhibitor | | Descriptor: | 3-{(E)-[(3-{[(2,6-dioxo-1,2,3,6-tetrahydropyrimidin-4-yl)methyl]amino}propoxy)imino]methyl}benzoic acid, SODIUM ION, THIOCYANATE ION, ... | | Authors: | Bianchet, M.A, Chung, S, Parker, J.B, Amzel, L.M, Stivers, J.T. | | Deposit date: | 2008-11-21 | | Release date: | 2009-04-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Impact of linker strain and flexibility in the design of a fragment-based inhibitor
Nat.Chem.Biol., 5, 2009
|
|
6T49
 
 | |
1USN
 
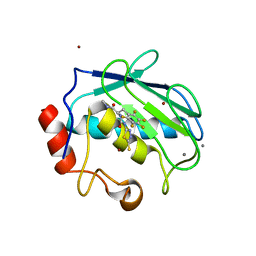 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN FIBROBLAST STROMELYSIN-1 INHIBITED WITH THIADIAZOLE INHIBITOR PNU-142372 | | Descriptor: | 2-[3-(5-MERCAPTO-[1,3,4]THIADIAZOL-2YL)-UREIDO]-N-METHYL-3-PENTAFLUOROPHENYL-PROPIONAMIDE, CALCIUM ION, STROMELYSIN-1, ... | | Authors: | Finzel, B.C, Bryant Junior, G.L, Baldwin, E.T. | | Deposit date: | 1998-06-09 | | Release date: | 1998-12-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterizations of nonpeptidic thiadiazole inhibitors of matrix metalloproteinases reveal the basis for stromelysin selectivity.
Protein Sci., 7, 1998
|
|
5O20
 
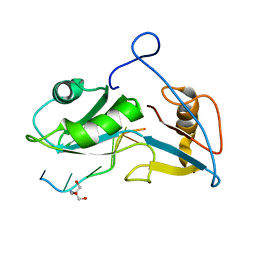 | | Structure of Nrd1 RNA binding domain in complex with RNA (UUAGUAAUCC) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Protein NRD1, RNA (5'-R(*UP*AP*GP*UP*AP*AP*UP*C)-3') | | Authors: | Franco-Echevarria, E, Perez-Canadillas, J.M, Gonzalez, B. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.53 Å) | | Cite: | The structure of transcription termination factor Nrd1 reveals an original mode for GUAA recognition.
Nucleic Acids Res., 45, 2017
|
|
5O2Y
 
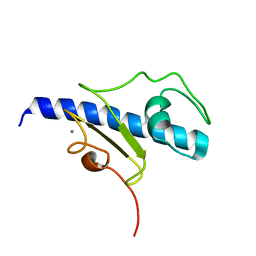 | | NMR structure of the calcium bound form of PulG, major pseudopilin from Klebsiella oxytoca T2SS | | Descriptor: | CALCIUM ION, General secretion pathway protein G | | Authors: | Lopez-Castilla, A, Bardiaux, B, Vitorge, B, Thomassin, J.-L, Zheng, W, Yu, X, Egelman, E.H, Nilges, M, Francetic, O, Izadi-Pruneyre, N. | | Deposit date: | 2017-05-23 | | Release date: | 2017-10-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the calcium-dependent type 2 secretion pseudopilus.
Nat Microbiol, 2, 2017
|
|
3FGS
 
 | |
7KR5
 
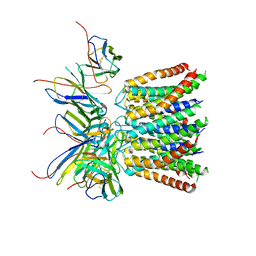 | | Cryo-EM structure of the CRAC channel Orai in an open conformation; H206A gain-of-function mutation in complex with an antibody | | Descriptor: | 19B5 Fab heavy chain, 19B5 Fab light chain, CALCIUM ION, ... | | Authors: | Long, S.B, Hou, X, Outhwaite, I.R. | | Deposit date: | 2020-11-18 | | Release date: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the calcium release-activated calcium channel Orai in an open conformation.
Elife, 9, 2020
|
|
3FMG
 
 | | Structure of rotavirus outer capsid protein VP7 trimer in complex with a neutralizing Fab | | Descriptor: | CALCIUM ION, Fab of neutralizing antibody 4F8, heavy chain, ... | | Authors: | Aoki, S.T, Settembre, E.C, Trask, S.D, Greenberg, H.B, Harrison, S.C, Dormitzer, P.R. | | Deposit date: | 2008-12-22 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of rotavirus outer-layer protein VP7 bound with a neutralizing Fab.
Science, 324, 2009
|
|
3FP3
 
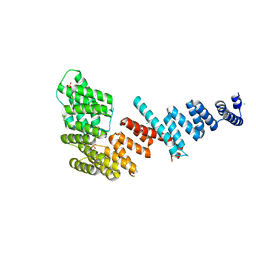 | | Crystal structure of Tom71 | | Descriptor: | CHLORIDE ION, SULFATE ION, TPR repeat-containing protein YHR117W | | Authors: | Li, J, Qian, X, Hu, J, Sha, B. | | Deposit date: | 2009-01-03 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Molecular chaperone Hsp70/Hsp90 prepares the mitochondrial outer membrane translocon receptor Tom71 for preprotein loading.
J.Biol.Chem., 284, 2009
|
|
7KX2
 
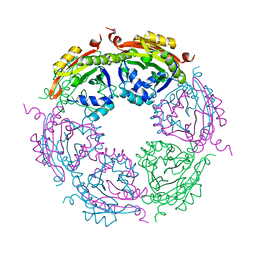 | | Spermidine N-acetyltransferase SpeG F149A mutant from Vibrio cholerae | | Descriptor: | Spermidine N(1)-acetyltransferase | | Authors: | Le, V.T.B, Tsimbalyuk, S, Lim, E.Q, Solis, A, Gawat, D, Boeck, P, Renolo, R, Forwood, J.K, Kuhn, M.L. | | Deposit date: | 2020-12-03 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Vibrio cholerae SpeG Spermidine/Spermine N -Acetyltransferase Allosteric Loop and beta 6-beta 7 Structural Elements Are Critical for Kinetic Activity.
Front Mol Biosci, 8, 2021
|
|
2LK1
 
 | |
5O66
 
 | | Asymmetric AcrABZ-TolC | | Descriptor: | Multidrug efflux pump accessory protein AcrZ, Multidrug efflux pump subunit AcrA, Multidrug efflux pump subunit AcrB, ... | | Authors: | Du, D, Luisi, B.F. | | Deposit date: | 2017-06-05 | | Release date: | 2017-06-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | An allosteric transport mechanism for the AcrAB-TolC multidrug efflux pump.
Elife, 6, 2017
|
|
2LNH
 
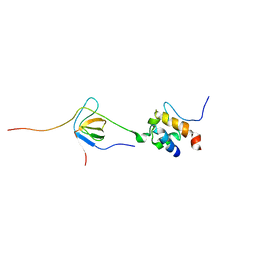 | | Enterohaemorrhagic E. coli (EHEC) exploits a tryptophan switch to hijack host F-actin assembly | | Descriptor: | Brain-specific angiogenesis inhibitor 1-associated protein 2-like protein 1, Neural Wiskott-Aldrich syndrome protein, Secreted effector protein EspF(U) | | Authors: | Aitio, O, Hellman, M, Skehan, B, Kesti, T, Leong, J.M, Saksela, K, Permi, P. | | Deposit date: | 2011-12-28 | | Release date: | 2012-08-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Enterohaemorrhagic Escherichia coli exploits a tryptophan switch to hijack host f-actin assembly.
Structure, 20, 2012
|
|
6TRW
 
 | |
3FDD
 
 | | The Crystal Structure of the Pseudomonas dacunhae Aspartate-Beta-Decarboxylase Reveals a Novel Oligomeric Assembly for a Pyridoxal-5-Phosphate Dependent Enzyme | | Descriptor: | ACETATE ION, CHLORIDE ION, L-aspartate-beta-decarboxylase, ... | | Authors: | Lima, S, Sundararaju, B, Huang, C, Khristoforov, R, Momany, C, Phillips, R.S. | | Deposit date: | 2008-11-25 | | Release date: | 2009-03-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The crystal structure of the Pseudomonas dacunhae aspartate-beta-decarboxylase dodecamer reveals an unknown oligomeric assembly for a pyridoxal-5'-phosphate-dependent enzyme.
J.Mol.Biol., 388, 2009
|
|
2LTD
 
 | | Solution NMR Structure of apo YdbC from Lactococcus lactis, Northeast Structural Genomics Consortium (NESG) Target KR150 | | Descriptor: | Uncharacterized protein ydbC | | Authors: | Rossi, P, Barbieri, C.M, Aramini, J.M, Bini, E, Lee, H, Janjua, H, Ciccosanti, C, Wang, H, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-05-16 | | Release date: | 2012-06-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structures of apo- and ssDNA-bound YdbC from Lactococcus lactis uncover the function of protein domain family DUF2128 and expand the single-stranded DNA-binding domain proteome.
Nucleic Acids Res., 41, 2013
|
|
1RRY
 
 | | DHNA complexed with 2-amino-4-hydroxy-5-carboxyethylpyrimidine | | Descriptor: | 2-AMINO-4-HYDROXYPYRIMIDINE-5-CARBOXYLIC ACID ETHYL ESTER, Dihydroneopterin aldolase | | Authors: | Sanders, W.J, Nienaber, V.L, Lerner, C.G, McCall, J.O, Merrick, S.M, Swanson, S.J, Harlan, J.E, Stoll, V.S, Stamper, G.F, Betz, S.F, Condroski, K.R, Meadows, R.P, Severin, J.M, Walter, K.A, Magdalinos, P, Jakob, C.G, Wagner, R, Beutel, B.A. | | Deposit date: | 2003-12-09 | | Release date: | 2004-03-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of Potent Inhibitors of Dihydroneopterin Aldolase Using CrystaLEAD High-Throughput X-ray Crystallographic Screening and Structure-Directed Lead Optimization.
J.Med.Chem., 47, 2004
|
|
5OE7
 
 | |
2L47
 
 | |
1RAU
 
 | |
