5LWH
 
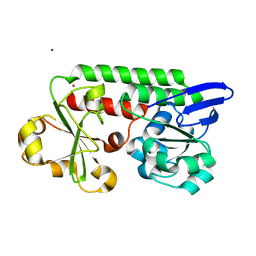 | | CeuE (Y288F variant) a periplasmic protein from Campylobacter jejuni. | | Descriptor: | Enterochelin ABC transporter substrate-binding protein, ZINC ION | | Authors: | Wilde, E.J, Blagova, E, Hughes, A, Raines, D.J, Moroz, O.V, Turkenburg, J, Duhme-Klair, A.-K, Wilson, K.S. | | Deposit date: | 2016-09-16 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Interactions of the periplasmic binding protein CeuE with Fe(III) n-LICAM(4-) siderophore analogues of varied linker length.
Sci Rep, 7, 2017
|
|
5LXB
 
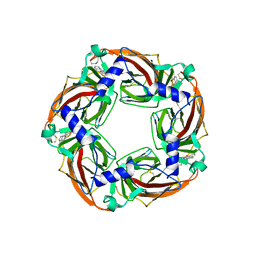 | |
5LZD
 
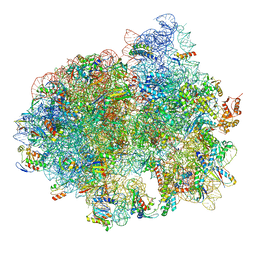 | | Structure of SelB-Sec-tRNASec bound to the 70S ribosome in the GTPase activated state (GA) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Bock, L.V, Maracci, C, Wang, Z, Paleskava, A, Konevega, A.L, Schroeder, G.F, Grubmueller, H, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The pathway to GTPase activation of elongation factor SelB on the ribosome.
Nature, 540, 2016
|
|
1FMO
 
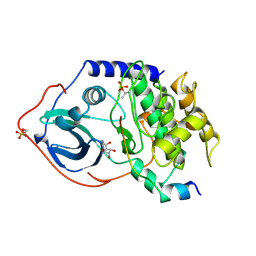 | | CRYSTAL STRUCTURE OF A POLYHISTIDINE-TAGGED RECOMBINANT CATALYTIC SUBUNIT OF CAMP-DEPENDENT PROTEIN KINASE COMPLEXED WITH THE PEPTIDE INHIBITOR PKI(5-24) AND ADENOSINE | | Descriptor: | ADENOSINE, CAMP-DEPENDENT PROTEIN KINASE, HEAT STABLE RABBIT SKELETAL MUSCLE INHIBITOR PROTEIN | | Authors: | Narayana, N, Cox, S, Shaltiel, S, Taylor, S.S, Xuong, N.-H. | | Deposit date: | 1997-07-08 | | Release date: | 1998-01-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a polyhistidine-tagged recombinant catalytic subunit of cAMP-dependent protein kinase complexed with the peptide inhibitor PKI(5-24) and adenosine.
Biochemistry, 36, 1997
|
|
5LZT
 
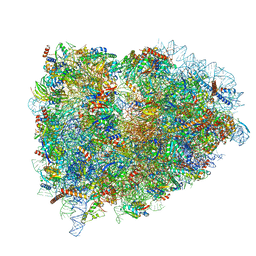 | | Structure of the mammalian ribosomal termination complex with eRF1 and eRF3. | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Shao, S, Murray, J, Brown, A, Taunton, J, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2016-10-02 | | Release date: | 2016-11-30 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Decoding Mammalian Ribosome-mRNA States by Translational GTPase Complexes.
Cell, 167, 2016
|
|
8TCE
 
 | |
5M36
 
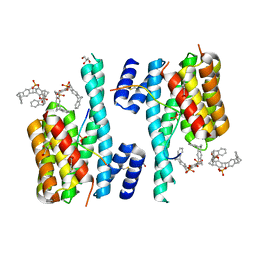 | | The molecular tweezer CLR01 stabilizes a disordered protein-protein interface | | Descriptor: | (1R,5S,9S,16R,20R,24S,28S,35R)-3,22-Bis(dihydroxyphosphoryloxy)tridecacyclo[22.14.1.15,20.19,16.128,35.02,23.04,21.06,19.08,17.010,15.025,38.027,36.029,34]dotetraconta-2(23),3,6,8(17),10,12,14,18,21,25,27(36),29,31,33,37-pentadecaene, 14-3-3 protein zeta/delta, BENZOIC ACID, ... | | Authors: | Bier, D, Ottmann, C. | | Deposit date: | 2016-10-14 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The Molecular Tweezer CLR01 Stabilizes a Disordered Protein-Protein Interface.
J. Am. Chem. Soc., 139, 2017
|
|
5M3K
 
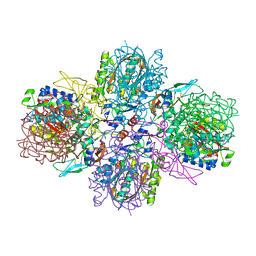 | | A multi-component acyltransferase PhlABC from Pseudomonas protegens | | Descriptor: | 2,4-diacetylphloroglucinol biosynthesis protein PhlB, 2,4-diacetylphloroglucinol biosynthesis protein PhlC, PhlA, ... | | Authors: | Pavkov-Keller, T, Schmidt, N.G, Kroutil, W, Gruber, K. | | Deposit date: | 2016-10-15 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structure and Catalytic Mechanism of a Bacterial Friedel-Crafts Acylase.
Chembiochem, 20, 2019
|
|
7TV4
 
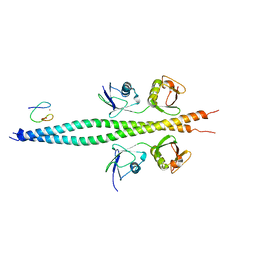 | | Crystal structure of NEMO CoZi in complex with HOIP NZF1 and linear diubiquitin | | Descriptor: | E3 ubiquitin-protein ligase RNF31, NF-kappa-B essential modulator, Polyubiquitin-C, ... | | Authors: | Rahighi, S, Iyer, M, Oveisi, H. | | Deposit date: | 2022-02-03 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Structural basis for the simultaneous recognition of NEMO and acceptor ubiquitin by the HOIP NZF1 domain.
Sci Rep, 12, 2022
|
|
6G1E
 
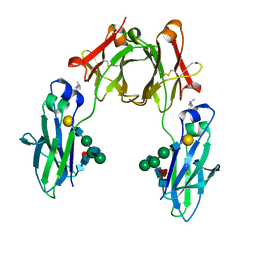 | | BEAT Fc with improved heterodimerization (Q3A-D84.4Q) | | Descriptor: | Immunoglobulin gamma-1 heavy chain, Immunoglobulin heavy constant gamma 1,Immunoglobulin heavy constant gamma 3, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Stutz, C, Blein, S. | | Deposit date: | 2018-03-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A single mutation increases heavy chain heterodimer assembly of bispecific antibodies by inducing structural disorder in one homodimer species.
J.Biol.Chem., 2020
|
|
6G20
 
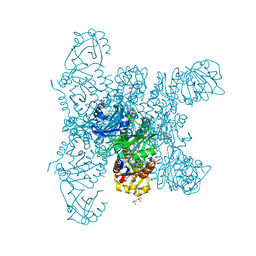 | | Crystal structure of a fluorescence optimized bathy phytochrome PAiRFP2 derived from wild-type Agp2 in its functional Meta-F intermediate state. | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3-[(2Z)-2-({3-(2-carboxyethyl)-5-[(E)-(4-ethenyl-3-methyl-5-oxo-1,5-dihydro-2H-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-2-yl}methylidene)-5-{(Z)-[(3E,4S)-3-ethylidene-4-methyl-5-oxopyrrolidin-2-ylidene]methyl}-4-methyl-2H-pyrrol-3-yl]propanoic acid, ... | | Authors: | Schmidt, A, Sauthof, L, Szczepek, M, Scheerer, P. | | Deposit date: | 2018-03-22 | | Release date: | 2018-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural snapshot of a bacterial phytochrome in its functional intermediate state.
Nat Commun, 9, 2018
|
|
6JRE
 
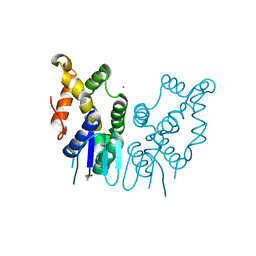 | | Structure of N-terminal domain of Plasmodium vivax p43 (PfNTD) solved by Co-SAD phasing | | Descriptor: | Aminoacyl-tRNA synthetase-interacting multifunctional protein p43, COBALT (II) ION | | Authors: | Manickam, Y, Harlos, K, Sharma, M, Gupta, S, Sharma, A. | | Deposit date: | 2019-04-03 | | Release date: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structures of the two domains that constitute the Plasmodium vivax p43 protein.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
7THN
 
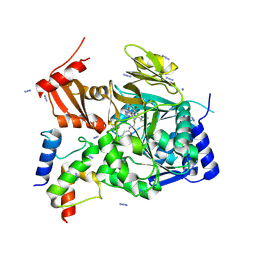 | |
6J5X
 
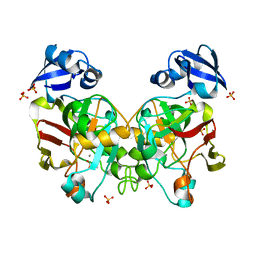 | | Crystal structure of fumarylpyruvate hydrolase from Corynebacterium glutamicum in complex with Mn2+ and pyruvate | | Descriptor: | MANGANESE (II) ION, PYRUVIC ACID, Predicted 2-keto-4-pentenoate hydratase/2-oxohepta-3-ene-1,7-dioic acid hydratase, ... | | Authors: | Hong, H, Seo, H, Kim, K.-J, Park, W. | | Deposit date: | 2019-01-12 | | Release date: | 2019-12-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Sequence, structure and function-based classification of the broadly conserved FAH superfamily reveals two distinct fumarylpyruvate hydrolase subfamilies.
Environ.Microbiol., 22, 2020
|
|
6JQS
 
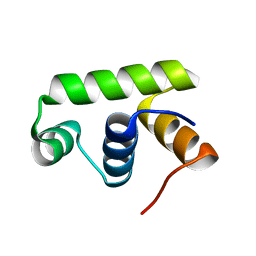 | | Structure of Transcription factor, GerE | | Descriptor: | DNA-binding response regulator | | Authors: | Lee, J.H, Lee, C.W. | | Deposit date: | 2019-04-01 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of a transcription factor, GerE (PaGerE), from spore-forming bacterium Paenisporosarcina sp. TG-14.
Biochem.Biophys.Res.Commun., 513, 2019
|
|
6J8V
 
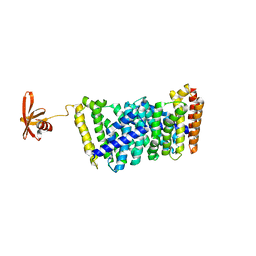 | | Structure of MOEN5-SSO7D fusion protein in complex with ligand 2 | | Descriptor: | FARNESYL, MoeN5,DNA-binding protein 7d | | Authors: | Ko, T.P, Zhang, L.L, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-01-21 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Complex structures of MoeN5 with substrate analogues suggest sequential catalytic mechanism.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
7U91
 
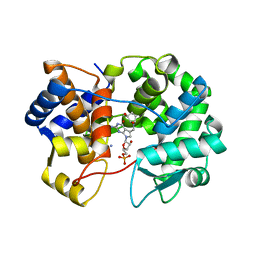 | | Crystal structure of queuine salvage enzyme DUF2419, in complex with queuosine-5'-monophosphate | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-7-(5-O-phosphono-beta-D-ribofuranosyl)-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, AMMONIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hung, S.-H, Swairjo, M.A. | | Deposit date: | 2022-03-09 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of Qng1-mediated salvage of the micronutrient queuine from queuosine-5'-monophosphate as the biological substrate.
Nucleic Acids Res., 51, 2023
|
|
5MW1
 
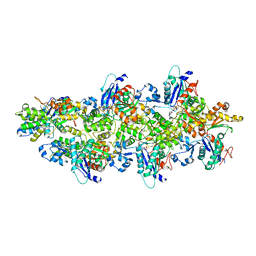 | |
7U07
 
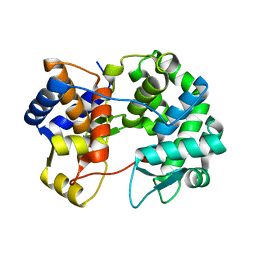 | |
7TUX
 
 | | Crystal Structure of Plasmodium falciparum Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferase in complex with [(3S)-4-Hydroxy-3-[({2-amino-4-hydroxy-5H-pyrrolo[3,2-d]pyrimidin-7-yl}methyl)amino]butyl]phosphonic acid | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Hypoxanthine-guanine-xanthine phosphoribosyltransferase, ... | | Authors: | Harijan, R.K, Minnow, Y.V.T, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2022-02-03 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Inhibition and Mechanism of Plasmodium falciparum Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferase.
Acs Chem.Biol., 17, 2022
|
|
5M63
 
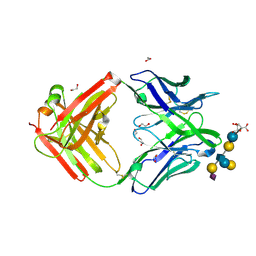 | | Crystal structure of group B Streptococcus type III DP2 oligosaccharide bound to Fab NVS-1-19-5 | | Descriptor: | (2~{R},3~{R},4~{S},5~{S})-2-[bis(oxidanyl)methyl]-5-(hydroxymethyl)oxolane-3,4-diol, 1,2-ETHANEDIOL, H chain of Fab NVS-1-19-5, ... | | Authors: | Carboni, F, Adamo, R, Veggi, D, Rappuoli, R, Malito, E, Margarit, I.R, Berti, F. | | Deposit date: | 2016-10-24 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structure of a protective epitope of group B Streptococcus type III capsular polysaccharide.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6G6U
 
 | | The dynamic nature of the VDAC1 channels in bilayers: human VDAC1 at 2.7 Angstrom resolution | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, NITRATE ION, Voltage-dependent anion-selective channel protein 1 | | Authors: | Razeto, A, Gribbon, P, Loew, C. | | Deposit date: | 2018-04-03 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | The dynamic nature of the VDAC1 channels in bilayers as revealed by two crystal structures of the human isoform in bicelles at 2.7 and 3.3 Angstrom resolution: implications for VDAC1 voltage-dependent mechanism and for its oligomerization
To Be Published
|
|
1FFP
 
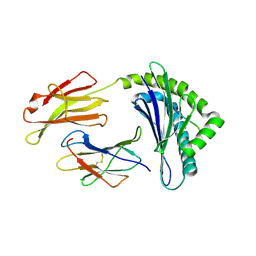 | | CRYSTAL STRUCTURE OF MURINE CLASS I H-2DB COMPLEXED WITH PEPTIDE GP33 (C9M/K1S) | | Descriptor: | BETA-2 MICROGLOBULIN BETA CHAIN, H-2 CLASS I HISTOCOMPATIBILITY ANTIGEN, D-B, ... | | Authors: | Wang, B, Sharma, A, Maile, R, Saad, M, Collins, E.J, Frelinger, J.A. | | Deposit date: | 2000-07-25 | | Release date: | 2002-12-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Peptidic termini play a significant role in TCR recognition
J.IMMUNOL., 169, 2002
|
|
1FG5
 
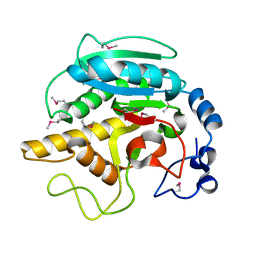 | | CRYSTAL STRUCTURE OF BOVINE ALPHA-1,3-GALACTOSYLTRANSFERASE CATALYTIC DOMAIN. | | Descriptor: | N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE | | Authors: | Gastinel, L.N, Bignon, C, Shaper, J.H, Joziasse, D.H. | | Deposit date: | 2000-07-28 | | Release date: | 2001-07-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Bovine alpha1,3-galactosyltransferase catalytic domain structure and its relationship with ABO histo-blood group and glycosphingolipid glycosyltransferases.
EMBO J., 20, 2001
|
|
7U1O
 
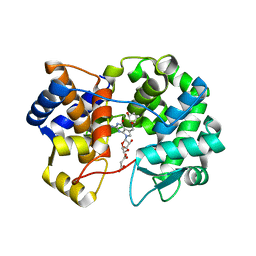 | | Crystal structure of queuine salvage enzyme DUF2419 complexed with queuosine | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-7-beta-D-ribofuranosyl-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, DI(HYDROXYETHYL)ETHER, MALONATE ION, ... | | Authors: | Hung, S.-H, Swairjo, M.A. | | Deposit date: | 2022-02-21 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis of Qng1-mediated salvage of the micronutrient queuine from queuosine-5'-monophosphate as the biological substrate.
Nucleic Acids Res., 51, 2023
|
|
