6IPA
 
 | |
5NON
 
 | | Structure of truncated Norcoclaurine Synthase from Thalictrum flavum with product mimic | | Descriptor: | 4-[2-[2-(4-methoxyphenyl)ethylamino]ethyl]benzene-1,2-diol, S-norcoclaurine synthase | | Authors: | Sula, A, Lichman, B.R, Pesnot, T, Ward, J.M, Hailes, H.C, Keep, N.H. | | Deposit date: | 2017-04-12 | | Release date: | 2017-09-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Evidence for the Dopamine-First Mechanism of Norcoclaurine Synthase.
Biochemistry, 56, 2017
|
|
5NOS
 
 | | Structure of cyclophilin A in complex with 3-amino-1H-pyridin-2-one | | Descriptor: | 5-azanyl-3~{H}-pyridin-6-one, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Georgiou, C, Mcnae, I.W, Ioannidis, H, Julien, M, Walkinshaw, M.D. | | Deposit date: | 2017-04-13 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Pushing the Limits of Detection of Weak Binding Using Fragment-Based Drug Discovery: Identification of New Cyclophilin Binders.
J. Mol. Biol., 429, 2017
|
|
6IPW
 
 | | Crystal structure of CqsB2 from Streptomyces exfoliatus in complex with the product, 1-(2-hydroxypropyl)-2-methyl-carbazole-3,4-dione | | Descriptor: | 2-methyl-1-[(2R)-2-oxidanylpropyl]-9H-carbazole-3,4-dione, CqsB2 | | Authors: | Tomita, T, Kobayashi, M, Nishiyama, M, Kuzuyama, T. | | Deposit date: | 2018-11-05 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An Unprecedented Cyclization Mechanism in the Biosynthesis of Carbazole Alkaloids in Streptomyces.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
5NQA
 
 | | Crystal structure of GalNAc-T4 in complex with the monoglycopeptide 3 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-galactopyranose, GLYCEROL, ... | | Authors: | de las Rivas, M, Lira-Navarrete, E, Daniel, E.J.P, Companon, I, Coelho, H, Diniz, A, Jimenez-Barbero, J, Peregrina, J.M, Clausen, H, Corzana, F, Marcelo, F, Jimenez-Oses, G, Gerken, T.A, Hurtado-Guerrero, R. | | Deposit date: | 2017-04-19 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The interdomain flexible linker of the polypeptide GalNAc transferases dictates their long-range glycosylation preferences.
Nat Commun, 8, 2017
|
|
6IFS
 
 | | KsgA from Bacillus subtilis 168 | | Descriptor: | Ribosomal RNA small subunit methyltransferase A | | Authors: | Bhujbalrao, R, Anand, R. | | Deposit date: | 2018-09-21 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Deciphering Determinants in Ribosomal Methyltransferases That Confer Antimicrobial Resistance.
J. Am. Chem. Soc., 141, 2019
|
|
5NRM
 
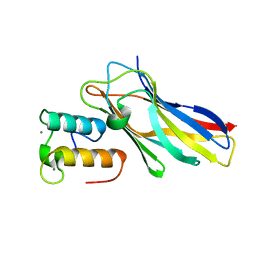 | | Crystal structure of the sixth cohesin from Acetivibrio cellulolyticus' scaffoldin B in complex with Cel5 dockerin S51I, L52N mutant | | Descriptor: | CALCIUM ION, DocCel5: Type I dockerin repeat domain from A. cellulolyticus family 5 endoglucanase WP_010249057 S51I, L52N mutant, ... | | Authors: | Bule, P, Najmudin, S, Fontes, C.M.G.A, Alves, V.D. | | Deposit date: | 2017-04-24 | | Release date: | 2018-01-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-function analyses generate novel specificities to assemble the components of multienzyme bacterial cellulosome complexes.
J. Biol. Chem., 293, 2018
|
|
5KUU
 
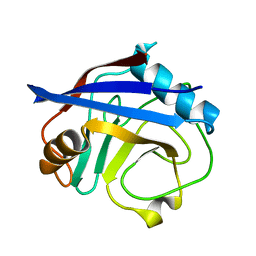 | | Human cyclophilin A at 100K, Data set 7 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Russi, S, Gonzalez, A, Kenner, L.R, Keedy, D.A, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2016-07-13 | | Release date: | 2016-08-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conformational variation of proteins at room temperature is not dominated by radiation damage.
J Synchrotron Radiat, 24, 2017
|
|
5KV2
 
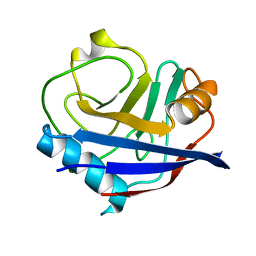 | | Human cyclophilin A at 278K, Data set 4 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Russi, S, Gonzalez, A, Kenner, L.R, Keedy, D.A, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2016-07-13 | | Release date: | 2016-08-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conformational variation of proteins at room temperature is not dominated by radiation damage.
J Synchrotron Radiat, 24, 2017
|
|
1FGX
 
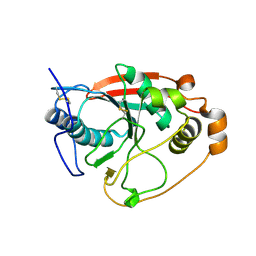 | | CRYSTAL STRUCTURE OF THE BOVINE BETA 1,4 GALACTOSYLTRANSFERASE (B4GALT1) CATALYTIC DOMAIN COMPLEXED WITH UMP | | Descriptor: | BETA 1,4 GALACTOSYLTRANSFERASE, URIDINE-5'-MONOPHOSPHATE | | Authors: | Gastinel, L.N, Cambillau, C, Bourne, Y. | | Deposit date: | 2000-07-29 | | Release date: | 2000-08-16 | | Last modified: | 2021-07-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the bovine beta4galactosyltransferase catalytic domain and its complex with uridine diphosphogalactose.
EMBO J., 18, 1999
|
|
5KYG
 
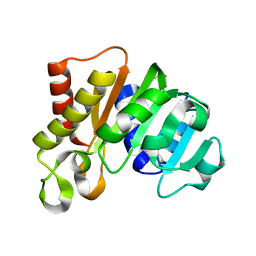 | |
1FIO
 
 | | CRYSTAL STRUCTURE OF YEAST T-SNARE PROTEIN SSO1 | | Descriptor: | SSO1 PROTEIN, ZINC ION | | Authors: | Munson, M, Chen, X, Cocina, A.E, Schultz, S.M, Hughson, F.M. | | Deposit date: | 2000-08-04 | | Release date: | 2000-10-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Interactions within the yeast t-SNARE Sso1p that control SNARE complex assembly.
Nat.Struct.Biol., 7, 2000
|
|
5N8E
 
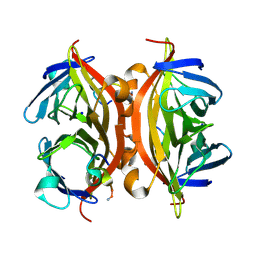 | | CRYSTAL STRUCTURE OF STREPTAVIDIN WITH PEPTIDE RDPAPAWAHGGG | | Descriptor: | ARG-ASP-PRO-ALA-PRO-ALA-TRP-ALA-HIS-GLY-GLY-GLY-NH2, SODIUM ION, Streptavidin | | Authors: | Lyamichev, V, Goodrich, L, Sullivan, E, Bannen, R, Benz, J, Albert, T, Patel, J. | | Deposit date: | 2017-02-23 | | Release date: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Stepwise Evolution Improves Identification of Diverse Peptides Binding to a Protein Target.
Sci Rep, 7, 2017
|
|
5N8X
 
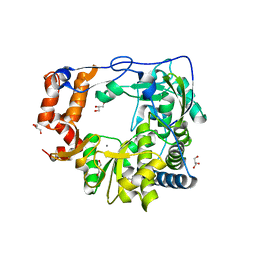 | |
5N9R
 
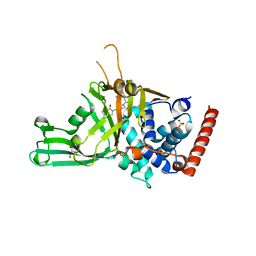 | | Crystal structure of USP7 in complex with a potent, selective and reversible small-molecule inhibitor | | Descriptor: | 7-bromanyl-3-[[4-oxidanyl-1-[(3~{R})-3-phenylbutanoyl]piperidin-4-yl]methyl]thieno[3,2-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Harrison, T, Gavory, G, O'Dowd, C, Helm, M, Flasz, I, Arkoudis, E, Dossang, A, Hughes, C, Cassidy, E, McClelland, K, Odrzywol, E, Page, N, Barker, O, Miel, H. | | Deposit date: | 2017-02-27 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Discovery and characterization of highly potent and selective allosteric USP7 inhibitors.
Nat. Chem. Biol., 14, 2018
|
|
1FL3
 
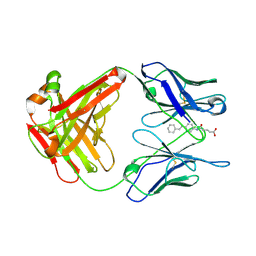 | |
5VD6
 
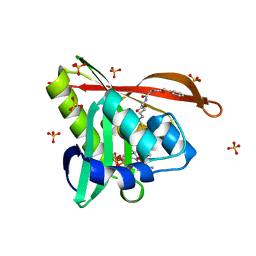 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with bisubstrate analog 6 | | Descriptor: | (3R,5S,9R,23S)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-10,14-dioxo-23-({[(phenylacetyl)amino]acetyl}amino)-2,4,6-trioxa-18-thia-11,15-diaza-3,5-diphosphatetracosan-24-oic acid 3,5-dioxide (non-preferred name), SULFATE ION, acetyltransferase PA4794 | | Authors: | Majorek, K.A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-04-01 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Generating enzyme and radical-mediated bisubstrates as tools for investigating Gcn5-related N-acetyltransferases.
FEBS Lett., 591, 2017
|
|
6IBS
 
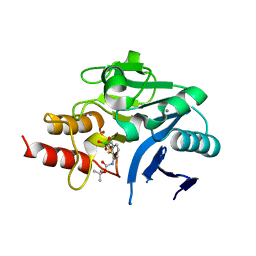 | | Crystal structure of NDM-1 beta-lactamase in complex with boronic inhibitor cpd 6 | | Descriptor: | CALCIUM ION, Metallo-beta-lactamase type 2, ZINC ION, ... | | Authors: | Maso, L, Quotadamo, A, Bellio, P, Montanari, M, Venturelli, A, Celenza, G, Costi, M.P, Tondi, D, Cendron, L. | | Deposit date: | 2018-11-30 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | X-ray Crystallography Deciphers the Activity of Broad-Spectrum Boronic Acid beta-Lactamase Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
5NGY
 
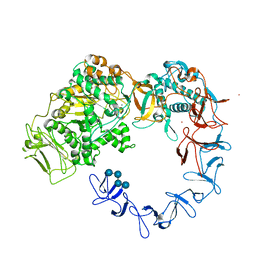 | | Crystal structure of Leuconostoc citreum NRRL B-1299 dextransucrase DSR-M | | Descriptor: | CALCIUM ION, DSR-M glucansucrase inactive mutant E715Q, PRASEODYMIUM ION, ... | | Authors: | Claverie, M, Cioci, G, Remaud-simeon, M, Moulis, C, Tranier, S, Vuillemin, M. | | Deposit date: | 2017-03-20 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Investigations on the Determinants Responsible for Low Molar Mass Dextran Formation by DSR-M Dextransucrase
Acs Catalysis, 2017
|
|
5KXK
 
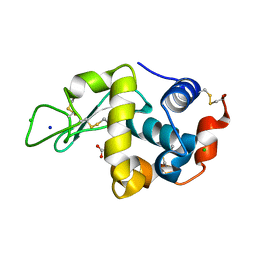 | | Hen Egg White Lysozyme at 100K, Data set 1 | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Russi, S, Gonzalez, A, Kenner, L.R, Keedy, D.A, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2016-07-20 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.198 Å) | | Cite: | Conformational variation of proteins at room temperature is not dominated by radiation damage.
J Synchrotron Radiat, 24, 2017
|
|
5L42
 
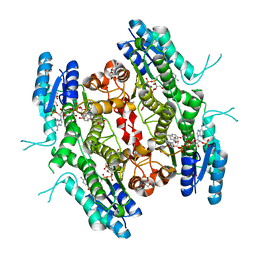 | | Leishmania major Pteridine reductase 1 (PTR1) in complex with compound 3 | | Descriptor: | (2~{R})-2-[3,4-bis(oxidanyl)phenyl]-6-oxidanyl-2,3-dihydrochromen-4-one, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Dello Iacono, L, Di Pisa, F, Pozzi, C, Landi, G, Mangani, S. | | Deposit date: | 2016-05-24 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Chroman-4-One Derivatives Targeting Pteridine Reductase 1 and Showing Anti-Parasitic Activity.
Molecules, 22, 2017
|
|
6I1H
 
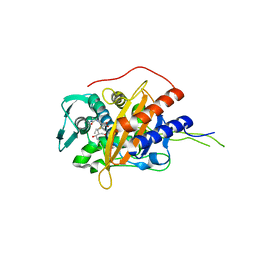 | | Crystal structure of TP domain from Chlamydia trachomatis Penicillin-Binding Protein 3 in complex with meropenem | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, Penicillin-binding protein,Penicillin-binding protein | | Authors: | Bellini, D, Koekemoer, L, Newman, H, Dowson, C.G. | | Deposit date: | 2018-10-28 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of TP domain from Chlamydia trachomatis Penicillin-Binding Protein 3 in complex with meropenem
To Be Published
|
|
5L00
 
 | | Self-complimentary RNA 15mer binding with GMP monomers | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, RNA (5'-R(*(LCC)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*GP*G)-3') | | Authors: | Zhang, W, Tam, C.P, Szostak, J.W. | | Deposit date: | 2016-07-26 | | Release date: | 2016-12-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Unusual Base-Pairing Interactions in Monomer-Template Complexes.
ACS Cent Sci, 2, 2016
|
|
6I1X
 
 | | Aeromonas hydrophila ExeD | | Descriptor: | Type II secretion system protein D | | Authors: | Contreras-Martel, C, Farias Estrozi, L. | | Deposit date: | 2018-10-30 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure and assembly of pilotin-dependent and -independent secretins of the type II secretion system.
Plos Pathog., 15, 2019
|
|
5L05
 
 | |
