4JXR
 
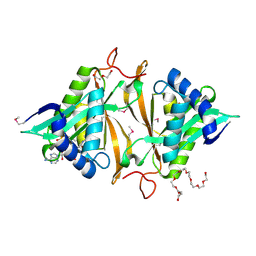 | | Crystal structure of a GNAT superfamily phosphinothricin acetyltransferase (Pat) from Sinorhizobium meliloti in complex with AcCoA | | Descriptor: | ACETYL COENZYME *A, Acetyltransferase, CITRATE ANION, ... | | Authors: | Majorek, K.A, Cooper, D.R, Porebski, P.J, Konina, K, Ahmed, M, Stead, M, Hillerich, B, Seidel, R, Bonanno, J.B, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-03-28 | | Release date: | 2013-05-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structure of a GNAT superfamily phosphinothricin acetyltransferase (Pat) from Sinorhizobium meliloti in complex with AcCoA
To be Published
|
|
5SUR
 
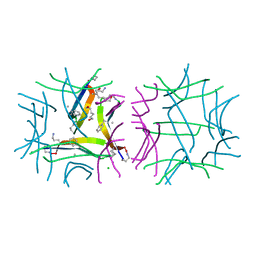 | | X-ray crystallographic structure of a covalent trimer derived from A-beta 17_36. Synchrotron data set. (ORN)CVF(MEA)CED(ORN)AIIGL(ORN)V. | | Descriptor: | 16mer A-beta peptide: ORN-CYS-VAL-PHE-MEA-CYS-GLU-ASP-ORN-ALA-ILE-ILE-GLY-LEU-ORN-VAL, CHLORIDE ION, HEXANE-1,6-DIOL, ... | | Authors: | Kreutzer, A.G, Yoo, S, Nowick, J.S. | | Deposit date: | 2016-08-03 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Stabilization, Assembly, and Toxicity of Trimers Derived from A beta.
J.Am.Chem.Soc., 139, 2017
|
|
5UIG
 
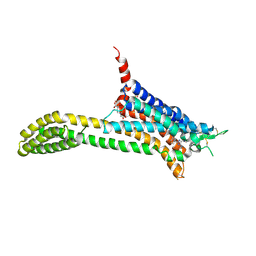 | | Crystal structure of adenosine A2A receptor bound to a novel triazole-carboximidamide antagonist | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-amino-N-[(2-methoxyphenyl)methyl]-2-(3-methylphenyl)-2H-1,2,3-triazole-4-carboximidamide, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, ... | | Authors: | Sun, B, Bachhawat, P, Ling-Hon Chu, M, Ceska, T, Sands, Z, Lebon, F, Kobilka, T.S, Kobilka, B. | | Deposit date: | 2017-01-13 | | Release date: | 2017-02-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the adenosine A2A receptor bound to an antagonist reveals a potential allosteric pocket.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7KPS
 
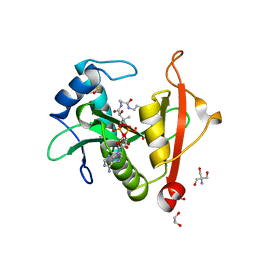 | | Structure of a GNAT superfamily PA3944 acetyltransferase in complex with AcCoA | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETYL COENZYME *A, ... | | Authors: | Czub, M.P, Porebski, P.J, Cymborowski, M, Reidl, C.T, Becker, D.P, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-12 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Gcn5-Related N- Acetyltransferases (GNATs) With a Catalytic Serine Residue Can Play Ping-Pong Too.
Front Mol Biosci, 8, 2021
|
|
4QQD
 
 | | Crystal Structure of tandem tudor domains of UHRF1 in complex with a small organic molecule | | Descriptor: | 4-methyl-2,3,4,5,6,7-hexahydrodicyclopenta[b,e]pyridin-8(1H)-imine, E3 ubiquitin-protein ligase UHRF1, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Iqbal, A, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-27 | | Release date: | 2014-08-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | A small molecule antagonist of SMN disrupts the interaction between SMN and RNAP II.
Nat Commun, 13, 2022
|
|
3IGJ
 
 | | Crystal Structure of Maltose O-acetyltransferase Complexed with Acetyl Coenzyme A from Bacillus anthracis | | Descriptor: | ACETYL COENZYME *A, FORMIC ACID, GLYCEROL, ... | | Authors: | Maltseva, N, Kim, Y, Papazisi, L, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-27 | | Release date: | 2009-08-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Maltose O-acetyltransferase Complexed with Acetyl Coenzyme A from Bacillus anthracis
To be Published
|
|
4EIQ
 
 | | Chromopyrrolic acid-soaked RebC-10x with bound 7-carboxy-K252c | | Descriptor: | (5S)-7-oxo-6,7,12,13-tetrahydro-5H-indolo[2,3-a]pyrrolo[3,4-c]carbazole-5-carboxylic acid, Putative FAD-monooxygenase | | Authors: | Goldman, P.J, Ryan, K.S, Howard-Jones, A.R, Hamill, M.J, Elliott, S.J, Walsh, C.T, Drennan, C.L. | | Deposit date: | 2012-04-05 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | An Unusual Role for a Mobile Flavin in StaC-like Indolocarbazole Biosynthetic Enzymes.
Chem.Biol., 19, 2012
|
|
1TT5
 
 | | Structure of APPBP1-UBA3-Ubc12N26: a unique E1-E2 interaction required for optimal conjugation of the ubiquitin-like protein NEDD8 | | Descriptor: | Ubiquitin-conjugating enzyme E2 M, ZINC ION, amyloid protein-binding protein 1, ... | | Authors: | Huang, D.T, Miller, D.W, Mathew, R, Cassell, R, Holton, J.M, Roussel, M.F, Schulman, B.A. | | Deposit date: | 2004-06-21 | | Release date: | 2004-09-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A unique E1-E2 interaction required for optimal conjugation of the ubiquitin-like protein NEDD8.
Nat.Struct.Mol.Biol., 11, 2004
|
|
3U6T
 
 | | Crystal structure of the complex of type I Ribosome inactivating protein in complex with Kanamycin at 1.85 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, KANAMYCIN A, ... | | Authors: | Yamini, S, Pandey, S, Kushwaha, G.S, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-10-13 | | Release date: | 2011-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the complex of type I Ribosome inactivating protein in complex with Kanamycin at 1.85 A
To be Published
|
|
5DOS
 
 | | Aurora A Kinase in Complex with AA35 and ATP in Space Group P6122 | | Descriptor: | 2-(3-bromophenyl)-8-fluoroquinoline-4-carboxylic acid, ADENOSINE-5'-TRIPHOSPHATE, Aurora kinase A, ... | | Authors: | Janecek, M, Rossmann, M, Sharma, P, Emery, A, McKenzie, G.J, Huggins, D.J, Stockwell, S, Stokes, J.A, Almeida, E.G, Hardwick, B, Narvaez, A.J, Hyvonen, M, Spring, D.R, Venkitaraman, A.R. | | Deposit date: | 2015-09-11 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Allosteric modulation of AURKA kinase activity by a small-molecule inhibitor of its protein-protein interaction with TPX2.
Sci Rep, 6, 2016
|
|
5DRD
 
 | | Aurora A Kinase in Complex with ATP in Space Group P6122 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Aurora kinase A, MAGNESIUM ION | | Authors: | Janecek, M, Rossmann, M, Sharma, P, Emery, A, McKenzie, G.J, Huggins, D.J, Stockwell, S, Stokes, J.A, Almeida, E.G, Hardwick, B, Narvaez, A.J, Hyvonen, M, Spring, D.R, Venkitaraman, A.R. | | Deposit date: | 2015-09-15 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Allosteric modulation of AURKA kinase activity by a small-molecule inhibitor of its protein-protein interaction with TPX2.
Sci Rep, 6, 2016
|
|
1Z1G
 
 | | Crystal structure of a lambda integrase tetramer bound to a Holliday junction | | Descriptor: | 25-MER, 29-MER, 5'-D(*AP*CP*AP*GP*GP*TP*CP*AP*CP*TP*AP*TP*CP*AP*GP*TP*CP*AP*AP*AP*AP*TP*AP*CP*C)-3', ... | | Authors: | Biswas, T, Aihara, H, Radman-Livaja, M, Filman, D, Landy, A, Ellenberger, T. | | Deposit date: | 2005-03-03 | | Release date: | 2005-06-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | A structural basis for allosteric control of DNA recombination by lambda integrase.
Nature, 435, 2005
|
|
5DT3
 
 | | Aurora A Kinase in Complex with ATP in Space Group P6122 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Aurora kinase A, MAGNESIUM ION, ... | | Authors: | Janecek, M, Rossmann, M, Sharma, P, Emery, A, McKenzie, G.J, Huggins, D.J, Stockwell, S, Stokes, J.A, Almeida, E.G, Hardwick, B, Narvaez, A.J, Hyvonen, M, Spring, D.R, Venkitaraman, A.R. | | Deposit date: | 2015-09-17 | | Release date: | 2016-07-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Allosteric modulation of AURKA kinase activity by a small-molecule inhibitor of its protein-protein interaction with TPX2.
Sci Rep, 6, 2016
|
|
4PZR
 
 | |
3SW2
 
 | | X-ray crystal structure of human FXA in complex with 6-chloro-N-((3S)-2-oxo-1-(2-oxo-2-((5S)-8-oxo-5,6-dihydro-1H-1,5-methanopyrido[1,2-a][1,5]diazocin-3(2H,4H,8H)-yl)ethyl)piperidin-3-yl)naphthalene-2-sulfonamide | | Descriptor: | 6-chloro-N-((3S)-2-oxo-1-(2-oxo-2-((5S)-8-oxo-5,6-dihydro-1H-1,5-methanopyrido[1,2-a][1,5]diazocin-3(2H,4H,8H)-yl)ethyl)piperidin-3-yl)naphthalene-2-sulfonamide, CALCIUM ION, Coagulation factor X, ... | | Authors: | Klei, H.E. | | Deposit date: | 2011-07-13 | | Release date: | 2011-11-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Arylsulfonamidopiperidone derivatives as a novel class of factor Xa inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
5WM1
 
 | | Structure of the 10S (+)-trans-BP-dG modified Rev1 ternary complex | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, 1,2-ETHANEDIOL, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Rechkoblit, O, Kolbanovsky, A, Landes, H, Geacintov, N.E, Aggarwal, A.K. | | Deposit date: | 2017-07-28 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of error-free replication across benzo[a]pyrene stereoisomers by Rev1 DNA polymerase.
Nat Commun, 8, 2017
|
|
4PZS
 
 | |
3H2G
 
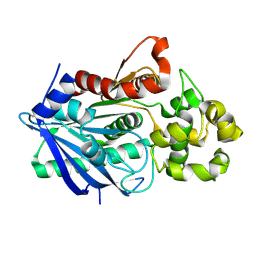 | |
3HTC
 
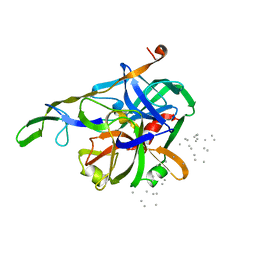 | | THE STRUCTURE OF A COMPLEX OF RECOMBINANT HIRUDIN AND HUMAN ALPHA-THROMBIN | | Descriptor: | ALPHA-THROMBIN (LARGE SUBUNIT), ALPHA-THROMBIN (SMALL SUBUNIT), HIRUDIN VARIANT 2 | | Authors: | Tulinsky, A, Rydel, T.J, Ravichandran, K.G, Huber, R, Bode, W. | | Deposit date: | 1993-06-11 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of a complex of recombinant hirudin and human alpha-thrombin.
Science, 249, 1990
|
|
4EIP
 
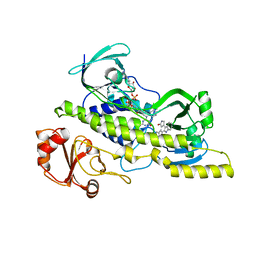 | | Native and K252c bound RebC-10x | | Descriptor: | 6,7,12,13-tetrahydro-5H-indolo[2,3-a]pyrrolo[3,4-c]carbazol-5-one, FLAVIN-ADENINE DINUCLEOTIDE, Putative FAD-monooxygenase | | Authors: | Goldman, P.J, Ryan, K.S, Howard-Jones, A.R, Hamill, M.J, Elliott, S.J, Walsh, C.T, Drennan, C.L. | | Deposit date: | 2012-04-05 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.332 Å) | | Cite: | An Unusual Role for a Mobile Flavin in StaC-like Indolocarbazole Biosynthetic Enzymes.
Chem.Biol., 19, 2012
|
|
3DTC
 
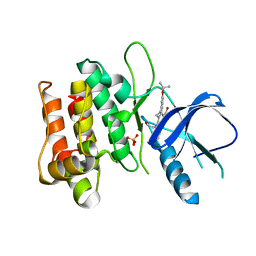 | | Crystal structure of mixed-lineage kinase MLK1 complexed with compound 16 | | Descriptor: | 12-(2-hydroxyethyl)-2-(1-methylethoxy)-13,14-dihydronaphtho[2,1-a]pyrrolo[3,4-c]carbazol-5(12H)-one, Mitogen-activated protein kinase kinase kinase 9, SULFATE ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Meyer, S.L, Hudkins, R.L, Almo, S.C. | | Deposit date: | 2008-07-14 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mixed-lineage kinase 1 and mixed-lineage kinase 3 subtype-selective dihydronaphthyl[3,4-a]pyrrolo[3,4-c]carbazole-5-ones: optimization, mixed-lineage kinase 1 crystallography, and oral in vivo activity in 1-methyl-4-phenyltetrahydropyridine models.
J.Med.Chem., 51, 2008
|
|
5BOO
 
 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM265 | | Descriptor: | 2-(1,1-difluoroethyl)-5-methyl-N-[4-(pentafluoro-lambda~6~-sulfanyl)phenyl][1,2,4]triazolo[1,5-a]pyrimidin-7-amine, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Phillips, M, Deng, X, Tomchick, D. | | Deposit date: | 2015-05-27 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A long-duration dihydroorotate dehydrogenase inhibitor (DSM265) for prevention and treatment of malaria.
Sci Transl Med, 7, 2015
|
|
5WM8
 
 | | Structure of the 10R (+)-cis-BP-dG modified Rev1 ternary complex | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, 1,2-ETHANEDIOL, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Rechkoblit, O, Kolbanovsky, A, Landes, H, Geacintov, N.E, Aggarwal, A.K. | | Deposit date: | 2017-07-28 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Mechanism of error-free replication across benzo[a]pyrene stereoisomers by Rev1 DNA polymerase.
Nat Commun, 8, 2017
|
|
3KZI
 
 | | Crystal Structure of Monomeric Form of Cyanobacterial Photosystem II | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Gabdulkhakov, A, Guskov, A, Broser, M, Kern, J, Zouni, A, Saenger, W. | | Deposit date: | 2009-12-08 | | Release date: | 2010-06-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal Structure of Monomeric Photosystem II from Thermosynechococcus elongatus at 3.6-A Resolution
J.Biol.Chem., 285, 2010
|
|
5EYC
 
 | | Crystal structure of c-Met in complex with naphthyridinone inhibitor 5 | | Descriptor: | 6-[(1~{R})-1-[8-fluoranyl-6-(3-methyl-1,2-oxazol-5-yl)-[1,2,4]triazolo[4,3-a]pyridin-3-yl]ethyl]-1,6-naphthyridin-5-one, Hepatocyte growth factor receptor | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2015-11-24 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of (R)-6-(1-(8-Fluoro-6-(1-methyl-1H-pyrazol-4-yl)-[1,2,4]triazolo[4,3-a]pyridin-3-yl)ethyl)-3-(2-methoxyethoxy)-1,6-naphthyridin-5(6H)-one (AMG 337), a Potent and Selective Inhibitor of MET with High Unbound Target Coverage and Robust In Vivo Antitumor Activity.
J.Med.Chem., 59, 2016
|
|
