8E9J
 
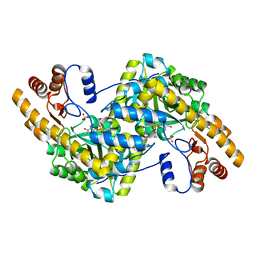 | | Crystal structure of E. coli aspartate aminotransferase mutant HEX in the ligand-free form at 278 K | | Descriptor: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8J0O
 
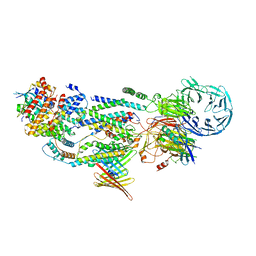 | | cryo-EM structure of human EMC and VDAC | | Descriptor: | ER membrane protein complex subunit 1, ER membrane protein complex subunit 10, ER membrane protein complex subunit 2, ... | | Authors: | Li, M, Zhang, C, Wu, J, Lei, M. | | Deposit date: | 2023-04-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structural insights into human EMC and its interaction with VDAC.
Aging (Albany NY), 16, 2024
|
|
8E9P
 
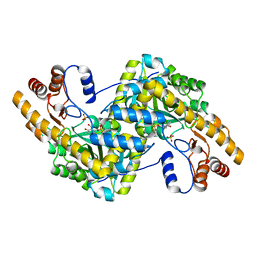 | | Crystal structure of wild-type E. coli aspartate aminotransferase in the ligand-free form at 278 K | | Descriptor: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8DU3
 
 | | Crystal structure of A2AAR-StaR2-bRIL in complex with compound 21a | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (4M)-6-bromo-4-(furan-2-yl)quinazolin-2-amine, Adenosine receptor A2a, ... | | Authors: | Shiriaeva, A, Stauch, B, Han, G.W, Cherezov, V. | | Deposit date: | 2022-07-26 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High ligand efficiency quinazoline compounds as novel A 2A adenosine receptor antagonists.
Eur.J.Med.Chem., 241, 2022
|
|
8E9D
 
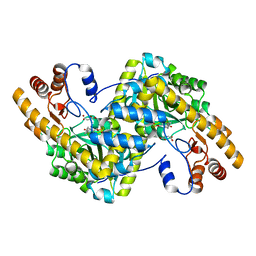 | | Crystal structure of E. coli aspartate aminotransferase mutant AIFS bound to maleic acid at 100 K | | Descriptor: | Aspartate aminotransferase, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8DT5
 
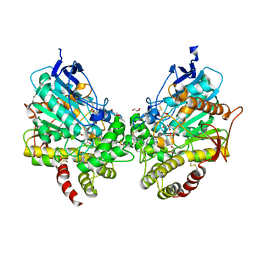 | | X-ray structure of human acetylcholinesterase ternary complex with paraoxon and oxime RS170B (POX-hAChE-RS170B) | | Descriptor: | 4-carbamoyl-1-(3-{2-[(E)-(hydroxyimino)methyl]-1H-imidazol-1-yl}propyl)pyridin-1-ium, Acetylcholinesterase, DIETHYL PHOSPHONATE, ... | | Authors: | Kovalevsky, A.Y, Gerlits, O, Radic, Z. | | Deposit date: | 2022-07-25 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and dynamic effects of paraoxon binding to human acetylcholinesterase by X-ray crystallography and inelastic neutron scattering.
Structure, 30, 2022
|
|
8IWN
 
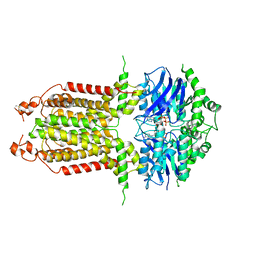 | | ABCG25 EQ mutant in ATP-bound state | | Descriptor: | ABC transporter G family member 25, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Sun, L, Liu, X, Ying, W, Liao, L, Wei, H. | | Deposit date: | 2023-03-30 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | ABCG25 EQ mutant in ATP-bound state
To Be Published
|
|
8E9C
 
 | | Crystal structure of E. coli aspartate aminotransferase mutant AIFS in the ligand-free form at 100 K | | Descriptor: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8J75
 
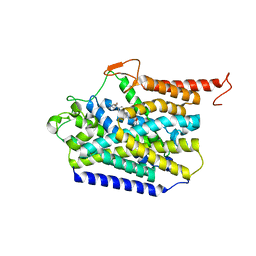 | | Human high-affinity choline transporter CHT1 in the HC-3-bound outward-facing open conformation, monomeric state | | Descriptor: | (2S,2'S)-2,2'-biphenyl-4,4'-diylbis(2-hydroxy-4,4-dimethylmorpholin-4-ium), High affinity choline transporter 1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Gao, Y, Qiu, Y, Zhao, Y. | | Deposit date: | 2023-04-27 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Transport mechanism of presynaptic high-affinity choline uptake by CHT1.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8J76
 
 | |
8E9U
 
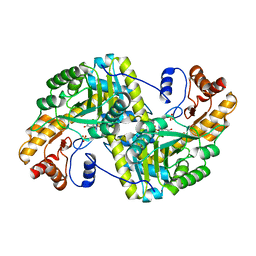 | | Crystal structure of E. coli aspartate aminotransferase mutant HEX in the ligand-free form at 303 K | | Descriptor: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8J2Z
 
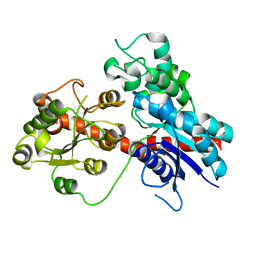 | |
5TJ8
 
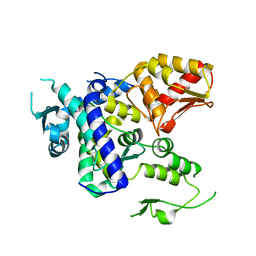 | |
8J2V
 
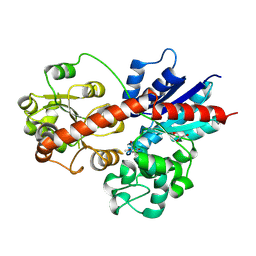 | |
8J2U
 
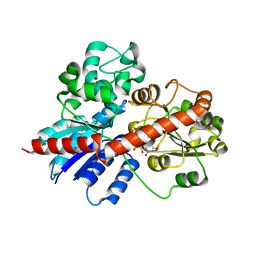 | |
5TAS
 
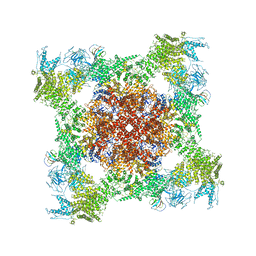 | | Structure of rabbit RyR1 (Caffeine/ATP/EGTA dataset, class 1) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, Peptidyl-prolyl cis-trans isomerase FKBP1B, ... | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-10 | | Release date: | 2016-10-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
8DYP
 
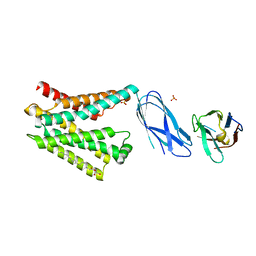 | |
8IWJ
 
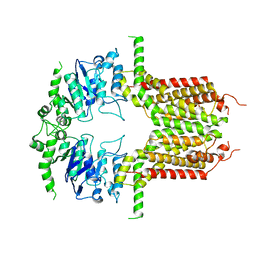 | |
8IWK
 
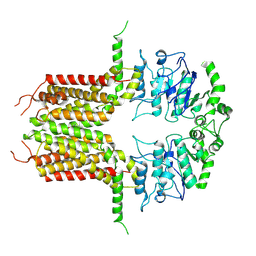 | | ABCG25 Wild Type purified with DDM plus CHS in ABA-bound state | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ABC transporter G family member 25 | | Authors: | Sun, L, Liu, X, Ying, W, Liao, L, Wei, H. | | Deposit date: | 2023-03-30 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | ABCG25 Wild Type purified with DDM plus CHS in ABA-bound state
To Be Published
|
|
8J31
 
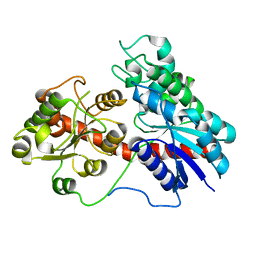 | |
8JJ3
 
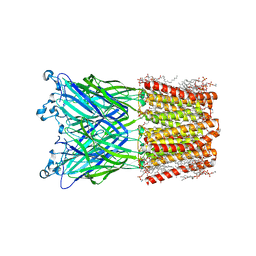 | | Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 2.5 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHLORIDE ION, Proton-gated ion channel | | Authors: | Bharambe, N, Li, Z, Basak, S. | | Deposit date: | 2023-05-29 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.6476 Å) | | Cite: | Cryo-EM structures of prokaryotic ligand-gated ion channel GLIC provide insights into gating in a lipid environment.
Nat Commun, 15, 2024
|
|
5TFC
 
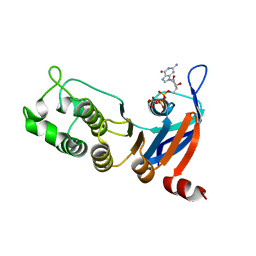 | | Nucleotide-binding domain 1 of the human cystic fibrosis transmembrane conductance regulator (CFTR) with GTP | | Descriptor: | Cystic fibrosis transmembrane conductance regulator, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Wang, C, Aleksandrov, A.A, Yang, Z, Forouhar, F, Proctor, E, Kota, P, An, J, Kaplan, A, Khazanov, N, Boel, G, Stockwell, B.R, Senderowitz, H, Dokholyan, N.V, Riordan, J.R, Brouillette, C.G, Hunt, J.F. | | Deposit date: | 2016-09-24 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Thermodynamic correction of F508del-CFTR by ligand binding to a remote site in the mutated domain
To Be Published
|
|
5TA3
 
 | | Structure of rabbit RyR1 (Caffeine/ATP/Ca2+ dataset, class 2) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-09 | | Release date: | 2016-10-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5TB2
 
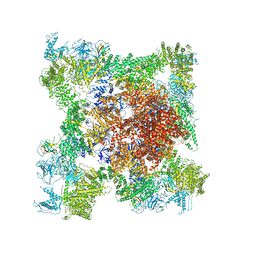 | | Structure of rabbit RyR1 (EGTA-only dataset, class 2) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ZINC ION | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-11 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
8DVP
 
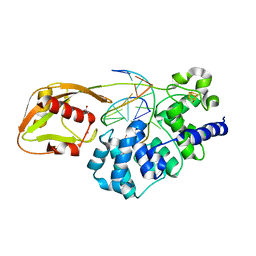 | | Glycosylase MutY variant N146S in complex with DNA containing d(8-oxo-G) paired with substrate purine | | Descriptor: | ACETATE ION, Adenine DNA glycosylase, CALCIUM ION, ... | | Authors: | Demir, M, Russelburg, L.P, Horvath, M.P, David, S.S. | | Deposit date: | 2022-07-29 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural snapshots of base excision by the cancer-associated variant MutY N146S reveal a retaining mechanism.
Nucleic Acids Res., 51, 2023
|
|
