4H9I
 
 | | Radiation damage study of lysozyme - 1.05 MGy | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C | | Authors: | Sutton, K.A, Snell, E.H. | | Deposit date: | 2012-09-24 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2002 Å) | | Cite: | Insights into the mechanism of X-ray-induced disulfide-bond cleavage in lysozyme crystals based on EPR, optical absorption and X-ray diffraction studies.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3ZQR
 
 | | NMePheB25 insulin analogue crystal structure | | Descriptor: | CHLORIDE ION, INSULIN A CHAIN, INSULIN B CHAIN, ... | | Authors: | Antolikova, E, Zakova, L, Turkenburg, J.P, Watson, C.J, Hanclova, I, Sanda, M, Cooper, A, Kraus, T, Brzozowski, A.M, Jiracek, J.A. | | Deposit date: | 2011-06-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Non-Equivalent Role of Inter- and Intramolecular Hydrogen Bonds in the Insulin Dimer Interface.
J.Biol.Chem., 286, 2011
|
|
4FVK
 
 | | Structural and functional characterization of neuraminidase-like molecule N10 derived from bat influenza A virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Li, Q, Sun, X.M, Li, Z.X, Liu, Y, Vavricka, C.J, Qi, J.X, Gao, G.F. | | Deposit date: | 2012-06-29 | | Release date: | 2012-09-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Structural and functional characterization of neuraminidase-like molecule N10 derived from bat influenza A virus
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3DH5
 
 | | Crystal structure of bovine pancreatic ribonuclease A (wild-type) | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic, SULFATE ION | | Authors: | Kurpiewska, K, Font, J, Ribo, M, Vilanova, M, Lewinski, K. | | Deposit date: | 2008-06-17 | | Release date: | 2008-07-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystallographic studies of RNase A variants engineered at the most destabilizing positions of the main hydrophobic core: further insight into protein stability
Proteins, 77, 2009
|
|
3ZKX
 
 | | TERNARY BACE2 XAPERONE COMPLEX | | Descriptor: | BETA-SECRETASE 2, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Kuglstatter, A, Banner, D.W, Benz, J, Bertschinger, J, Burger, D, Cuppuleri, S, Debulpaep, M, Gast, A, Grabulovski, D, Gsell, B, Hilpert, H, Huber, W, Kusznir, E, Laeremans, T, Matile, H, Rufer, A, Schlatter, D, Steyeart, J, Stihle, M, Thoma, R, Weber, M, Ruf, A. | | Deposit date: | 2013-01-25 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Mapping the Conformational Space Accessible to Bace2 Using Surface Mutants and Co-Crystals with Fab-Fragments, Fynomers, and Xaperones
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3DHR
 
 | | Crystal Structure Determination of Methemoglobin from Pigeon at 2 Angstrom Resolution (Columba livia) | | Descriptor: | Hemoglobin subunit alpha-A, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sathya Moorthy, P, Neelagandan, K, Balasubramanian, M, Ponnuswamy, M.N. | | Deposit date: | 2008-06-18 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Purification, Crystallization and X-ray Diffraction Studies of Pigeon Hemoglobin
To be Published
|
|
3DE7
 
 | |
3DE4
 
 | |
3ZBV
 
 | | Crystal Structure of murine Angiogenin-2 | | Descriptor: | ANGIOGENIN-2, GLYCEROL, SULFATE ION, ... | | Authors: | Iyer, S, Holloway, D.E, Acharya, K.R. | | Deposit date: | 2012-11-13 | | Release date: | 2012-12-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal Structures of Murine Angiogenin-2 and -3 - Probing 'Structure - Function' Relationships Amongst Angiogenin Homologues.
FEBS J., 280, 2013
|
|
4KE1
 
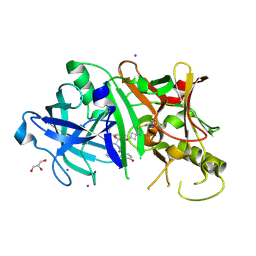 | | Crystal structure of BACE1 in complex with hydroxyethylamine-macrocyclic inhibitor 19 | | Descriptor: | (12S)-12-[(1R)-2-{[(4S)-6-ethyl-3,4-dihydrospiro[chromene-2,1'-cyclobutan]-4-yl]amino}-1-hydroxyethyl]-1,13-diazatricyclo[13.3.1.1~6,10~]icosa-6(20),7,9,15(19),16-pentaene-14,18-dione, Beta-Secretase 1, GLYCEROL, ... | | Authors: | Whittington, D.A, Long, A.M, Li, V. | | Deposit date: | 2013-04-25 | | Release date: | 2013-07-03 | | Last modified: | 2013-07-17 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Hydroxyethylamine-based inhibitors of BACE1: P1-P3 macrocyclization can improve potency, selectivity, and cell activity.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3DI7
 
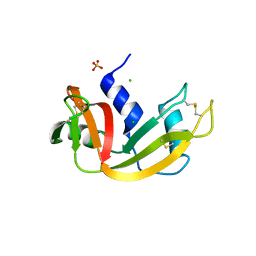 | | Crystal structure of bovine pancreatic ribonuclease A variant (V54A) | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic, SULFATE ION | | Authors: | Kurpiewska, K, Font, J, Ribo, M, Vilanova, M, Lewinski, K. | | Deposit date: | 2008-06-20 | | Release date: | 2008-07-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystallographic studies of RNase A variants engineered at the most destabilizing positions of the main hydrophobic core: further insight into protein stability
Proteins, 77, 2009
|
|
3DJX
 
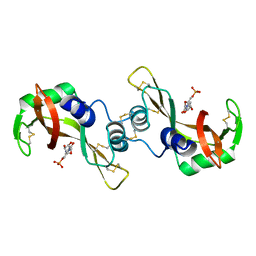 | | Bovine Seminal Ribonuclease- cytidine 5' phosphate complex | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, Seminal ribonuclease | | Authors: | Dossi, K, Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G. | | Deposit date: | 2008-06-24 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Mapping the ribonucleolytic active site of bovine seminal ribonuclease. The binding of pyrimidinyl phosphonucleotide inhibitors
Eur.J.Med.Chem., 44, 2009
|
|
3ZPQ
 
 | | Thermostabilised turkey beta1 adrenergic receptor with 4-(piperazin-1- yl)-1H-indole bound (compound 19) | | Descriptor: | 4-(PIPERAZIN-1-YL)-1H-INDOLE, BETA-1 ADRENERGIC RECEPTOR, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Christopher, J.A, Congreve, M, Dore, A.S, Marshall, F.H, Myszka, D.G, Brown, J, Koglin, M, Tehan, B, Errey, J.C, Tate, C.G, Warne, T. | | Deposit date: | 2013-03-01 | | Release date: | 2013-04-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Biophysical Fragment Screening of the Beta1-Adrenergic Receptor: Identification of High Affinity Aryl Piperazine Leads Using Structure-Based Drug Design.
J.Med.Chem., 56, 2013
|
|
3ZSN
 
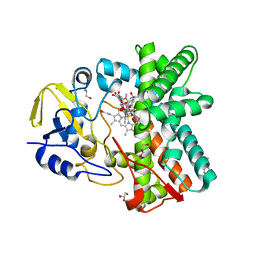 | | Structure of the mixed-function P450 MycG F286A mutant in complex with mycinamicin IV | | Descriptor: | BENZAMIDINE, GLYCEROL, MYCINAMICIN IV, ... | | Authors: | Li, S, Kells, P.M, Rutaganira, F.U, Anzai, Y, Kato, F, Sherman, D.H, Podust, L.M. | | Deposit date: | 2011-06-29 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate Recognition by the Multifunctional Cytochrome P450 Mycg in Mycinamicin Hydroxylation and Epoxidation Reactions.
J.Biol.Chem., 287, 2012
|
|
4FUH
 
 | | Crystal Structure of the Urokinase | | Descriptor: | 6-[(phenylcarbamoyl)amino]naphthalene-2-carboximidamide, ACETATE ION, GLYCEROL, ... | | Authors: | Kang, Y.N, Stuckey, J.A, Nienaber, V, Giranda, V. | | Deposit date: | 2012-06-28 | | Release date: | 2012-08-22 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Urokinase
to be published
|
|
3ZD2
 
 | | THE STRUCTURE OF THE TWO N-TERMINAL DOMAINS OF COMPLEMENT FACTOR H RELATED PROTEIN 1 SHOWS FORMATION OF A NOVEL DIMERISATION INTERFACE | | Descriptor: | 1,2-ETHANEDIOL, COMPLEMENT FACTOR H-RELATED PROTEIN 1 | | Authors: | Caesar, J.J.E, Goicoechea de Jorge, E, Pickering, M.C, Lea, S.M. | | Deposit date: | 2012-11-23 | | Release date: | 2013-03-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Dimerization of Complement Factor H-Related Proteins Modulates Complement Activation in Vivo.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3ZU1
 
 | |
3DR9
 
 | | Increased Distal Histidine Conformational Flexibility in the Deoxy Form of Dehaloperoxidase from Amphitrite ornata | | Descriptor: | Dehaloperoxidase A, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Chen, X, de Serrano, V.S, Betts, L, Franzen, S. | | Deposit date: | 2008-07-10 | | Release date: | 2009-01-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Distal histidine conformational flexibility in dehaloperoxidase from Amphitrite ornata.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3ZG3
 
 | | STEROL 14-ALPHA DEMETHYLASE (CYP51)FROM TRYPANOSOMA CRUZI IN COMPLEX WITH THE PYRIDINE INHIBITOR N-(1-(5-(trifluoromethyl)(pyridin-2-yl)) piperidin-4yl)-N-(4-(trifluoromethyl)phenyl)pyridin-3-amine (EPL- BS967, UDD) | | Descriptor: | N-[4-(trifluoromethyl)phenyl]-N-[1-[5-(trifluoromethyl)pyridin-2-yl]piperidin-4-yl]pyridin-3-amine, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE | | Authors: | Hargrove, T.Y, Wawrzak, Z, Keenan, M, Chatelain, E, Lepesheva, G.I. | | Deposit date: | 2012-12-14 | | Release date: | 2013-09-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Complexes of Trypanosoma Cruzi Sterol 14Alpha-Demethylase (Cyp51) with Two Pyridine-Based Drug Candidates for Chagas Disease: Structural Basis for Pathogen-Selectivity
J.Biol.Chem., 288, 2013
|
|
4GB2
 
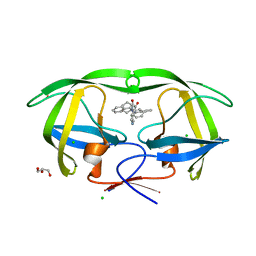 | | HIV-1 protease (mutant Q7K L33I L63I) in complex with a bicyclic pyrrolidine inhibitor | | Descriptor: | (4aS,7aS)-1,4-bis(diphenylmethyl)hexahydro-1H-pyrrolo[3,4-b]pyrazine-2,3-dione, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stieler, M, Heine, A, Klebe, G. | | Deposit date: | 2012-07-26 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.788 Å) | | Cite: | Cocrystallization of potent pyrrolidine based HIV-1 protease inhibitors
To be Published
|
|
4GC0
 
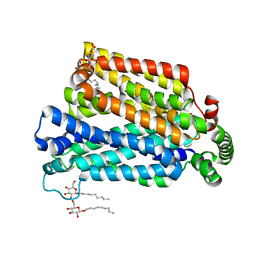 | | The structure of the MFS (major facilitator superfamily) proton:xylose symporter XylE bound to 6-bromo-6-deoxy-D-glucose | | Descriptor: | 6-bromo-6-deoxy-beta-D-glucopyranose, D-xylose-proton symporter, nonyl beta-D-glucopyranoside | | Authors: | Yan, N, Sun, L.F, Zeng, X, Yan, C.Y. | | Deposit date: | 2012-07-28 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a bacterial homologue of glucose transporters GLUT1-4.
Nature, 490, 2012
|
|
3DSK
 
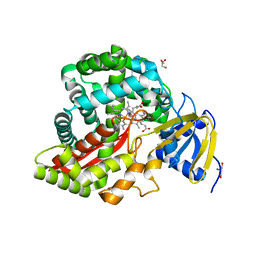 | | Crystal Structure of Arabidopsis thaliana Allene Oxide Synthase variant (F137L) (At-AOS(F137L), Cytochrome P450 74A, CYP74A) Complexed with 12R,13S-Vernolic Acid at 1.55 A Resolution | | Descriptor: | (9Z)-11-[(2R,3S)-3-pentyloxiran-2-yl]undec-9-enoic acid, Cytochrome P450 74A, chloroplast, ... | | Authors: | Lee, D.S, Nioche, P, Raman, C.S. | | Deposit date: | 2008-07-12 | | Release date: | 2008-08-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insights into the evolutionary paths of oxylipin biosynthetic enzymes
Nature, 455, 2008
|
|
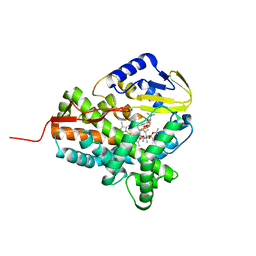
3ZKP
 
 | |
4GCH
 
 | |
4K45
 
 | |
