6RJN
 
 | | Crystal structure of a Fungal Catalase at 2.3 Angstroms | | Descriptor: | CHLORIDE ION, Catalase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gomez, S, Navas-Yuste, S, Payne, A.M, Rivera, W, Lopez-Estepa, M, Brangbour, C, Fulla, D, Juanhuix, J, Fernandez, F.J, Vega, M.C. | | Deposit date: | 2019-04-28 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Peroxisomal catalases from the yeasts Pichia pastoris and Kluyveromyces lactis as models for oxidative damage in higher eukaryotes.
Free Radic. Biol. Med., 141, 2019
|
|
2G50
 
 | | The location of the allosteric amino acid binding site of muscle pyruvate kinase. | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ALANINE, ... | | Authors: | Holyoak, T, Williams, R, Fenton, A.W. | | Deposit date: | 2006-02-22 | | Release date: | 2006-05-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Differentiating a Ligand's Chemical Requirements for Allosteric Interactions from Those for Protein Binding. Phenylalanine Inhibition of Pyruvate Kinase.
Biochemistry, 45, 2006
|
|
4NWC
 
 | | Crystal structure of the GluK3 ligand-binding domain (S1S2) in complex with the agonist (2S,4R)-4-(3-Methoxy-3-oxopropyl)glutamic acid at 2.01 A resolution. | | Descriptor: | (2S,4R)-4-(3-Methoxy-3-oxopropyl) glutamic acid, CHLORIDE ION, Glutamate receptor ionotropic, ... | | Authors: | Larsen, A.P, Venskutonyte, R, Frydenvang, K, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-12-06 | | Release date: | 2014-08-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.012 Å) | | Cite: | Molecular Recognition of Two 2,4-syn-Functionalized (S)-Glutamate Analogues by the Kainate Receptor GluK3 Ligand Binding Domain.
Chemmedchem, 9, 2014
|
|
2EZ2
 
 | | Apo tyrosine phenol-lyase from Citrobacter freundii at pH 8.0 | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, Tyrosine phenol-lyase | | Authors: | Milic, D, Matkovic-Calogovic, D, Demidkina, T.V, Antson, A.A. | | Deposit date: | 2005-11-10 | | Release date: | 2006-07-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of apo- and holo-tyrosine phenol-lyase reveal a catalytically critical closed conformation and suggest a mechanism for activation by K+ ions
Biochemistry, 45, 2006
|
|
2HIT
 
 | | Reaction centre from Rhodobacter sphaeroides strain R-26.1 complexed with dibrominated phosphatidylethanolamine | | Descriptor: | (1R)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (9S,10S)-9,10-DIBROMOOCTADECANOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, BACTERIOCHLOROPHYLL A, ... | | Authors: | Roszak, A.W, Gardiner, A.T, Isaacs, N.W, Cogdell, R.J. | | Deposit date: | 2006-06-29 | | Release date: | 2007-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Brominated Lipids Identify Lipid Binding Sites on the Surface of the Reaction Center from Rhodobacter sphaeroides.
Biochemistry, 46, 2007
|
|
2HHK
 
 | | Reaction centre from Rhodobacter sphaeroides strain R-26.1 complexed with dibrominated phosphatidylglycerol | | Descriptor: | (1R)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (9S,10S)-9,10-DIBROMOOCTADECANOATE, (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, BACTERIOCHLOROPHYLL A, ... | | Authors: | Roszak, A.W, Gardiner, A.T, Isaacs, N.W, Cogdell, R.J. | | Deposit date: | 2006-06-28 | | Release date: | 2007-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Brominated Lipids Identify Lipid Binding Sites on the Surface of the Reaction Center from Rhodobacter sphaeroides.
Biochemistry, 46, 2007
|
|
2HH1
 
 | | Reaction centre from Rhodobacter sphaeroides strain R-26.1 complexed with dibrominated phosphatidylcholine | | Descriptor: | (7R,14S)-14,15-DIBROMO-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, BACTERIOCHLOROPHYLL A, ... | | Authors: | Roszak, A.W, Gardiner, A.T, Isaacs, N.W, Cogdell, R.J. | | Deposit date: | 2006-06-27 | | Release date: | 2007-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Brominated Lipids Identify Lipid Binding Sites on the Surface of the Reaction Center from Rhodobacter sphaeroides.
Biochemistry, 46, 2007
|
|
1KQE
 
 | | Solution structure of a linked shortened gramicidin A in benzene/acetone 10:1 | | Descriptor: | MINI-GRAMICIDIN A | | Authors: | Arndt, H.D, Bockelmann, D, Knoll, A, Lamberth, S, Griesinger, C, Koert, U. | | Deposit date: | 2002-01-05 | | Release date: | 2002-11-27 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Cation Control in Functional Helical Programming: Structures of a D,L-Peptide Ion Channel
Angew.Chem.Int.Ed.Engl., 41, 2002
|
|
7XNY
 
 | |
7XNX
 
 | |
3MZ8
 
 | | Crystal Structure of Zinc-Bound Natrin From Naja atra | | Descriptor: | Natrin-1, ZINC ION | | Authors: | Wang, Y.L, Hsieh, Y.C, Liu, J.S, Chen, C.J, Wu, W.G. | | Deposit date: | 2010-05-12 | | Release date: | 2010-09-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cobra CRISP functions as an inflammatory modulator via a novel Zn2+- and heparan sulfate- dependent transcriptional regulation of endothelial cell adhesion molecules
J.Biol.Chem., 285, 2010
|
|
4HYV
 
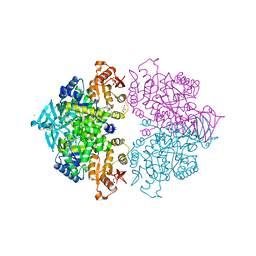 | | Pyruvate kinase (PYK) from Trypanosoma brucei in the presence of Magnesium, PEP and F26BP | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Zhong, W, Morgan, H.P, McNae, I.W, Michels, P.A.M, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2012-11-14 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | `In crystallo' substrate binding triggers major domain movements and reveals magnesium as a co-activator of Trypanosoma brucei pyruvate kinase.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HYW
 
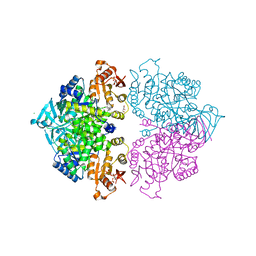 | | Pyruvate kinase (PYK) from Trypanosoma brucei in the presence of Magnesium and F26BP | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Zhong, W, Morgan, H.P, McNae, I.W, Michels, P.A.M, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2012-11-14 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | `In crystallo' substrate binding triggers major domain movements and reveals magnesium as a co-activator of Trypanosoma brucei pyruvate kinase.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1MGY
 
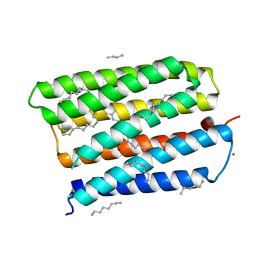 | | Structure of the D85S mutant of bacteriorhodopsin with bromide bound | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, BROMIDE ION, Bacteriorhodopsin, ... | | Authors: | Facciotti, M.T, Cheung, V.S, Nguyen, D, Rouhani, S, Glaeser, R.M. | | Deposit date: | 2002-08-16 | | Release date: | 2003-07-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Bromide-Bound D85S Mutant of Bacteriorhodopsin:
Principles of Ion Pumping
Biophys.J., 85, 2003
|
|
4IIN
 
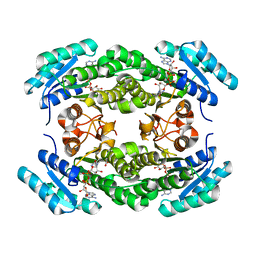 | | Crystal structure of a putative 3-oxoacyl-[acyl-carrier protein]reductase from Helicobacter pylori 26695 complexed with NAD+ | | Descriptor: | 3-ketoacyl-acyl carrier protein reductase (FabG), ACETATE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Hou, J, Osinski, T, Zheng, H, Shumilin, I, Shabalin, I, Shatsman, S, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-12-20 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a putative 3-oxoacyl-[acyl-carrier protein]reductase from Helicobacter pylori 26695 complexed with NAD+
To be Published
|
|
4G8N
 
 | | Crystal structure of the kainate receptor GluK3 ligand-binding domain in complex with the agonist G8M | | Descriptor: | (1S,2R)-2-[(S)-amino(carboxy)methyl]cyclobutanecarboxylic acid, CHLORIDE ION, Glutamate receptor, ... | | Authors: | Venskutonyte, R, Kastrup, J.S, Frydenvang, K, Gajhede, M. | | Deposit date: | 2012-07-23 | | Release date: | 2012-08-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Pharmacological and structural characterization of conformationally restricted (S)-glutamate analogues at ionotropic glutamate receptors.
J.Struct.Biol., 180, 2012
|
|
4E0W
 
 | | Crystal structure of the kainate receptor GluK3 ligand binding domain in complex with kainate | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, CHLORIDE ION, Glutamate receptor, ... | | Authors: | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2012-03-05 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3501 Å) | | Cite: | Kainate induces various domain closures in AMPA and kainate receptors.
Neurochem Int, 61, 2012
|
|
4KCT
 
 | | Pyruvate kinase (PYK) from Trypanosoma brucei soaked with Oxaloacetate | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Zhong, W, Morgan, H.P, McNae, I.W, Michels, P.A.M, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2013-04-24 | | Release date: | 2014-01-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pyruvate kinases have an intrinsic and conserved decarboxylase activity.
Biochem.J., 458, 2014
|
|
4IP0
 
 | | X-Ray Structure of the Complex Uridine Phosphorylase from Vibrio cholerae with Phosphate Ion at 1.29 A Resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, ETHANOL, ... | | Authors: | Prokofev, I.I, Lashkov, A.A, Gabdoulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2013-01-09 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.294 Å) | | Cite: | X-ray structures of uridine phosphorylase from Vibrio cholerae in complexes with uridine, thymidine, uracil, thymine, and phosphate anion: Substrate specificity of bacterial uridine phosphorylases
Crystallography Reports, 61, 2016
|
|
3I4D
 
 | | Photosynthetic reaction center from rhodobacter sphaeroides 2.4.1 | | Descriptor: | (2R,3S)-heptane-1,2,3-triol, 1,4-DIETHYLENE DIOXIDE, BACTERIOCHLOROPHYLL A, ... | | Authors: | Fujii, R, Adachi, S, Roszak, A.W, Gardiner, A.T, Cogdell, R.J, Isaacs, N.W, Koshihara, S, Hashimoto, H. | | Deposit date: | 2009-07-01 | | Release date: | 2010-12-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of the carotenoid bound to the reaction centre from Rhodobacter sphaeroides 2.4.1 revealed by time-resolved X-ray crystallography
To be Published
|
|
4KRZ
 
 | | Apo crystal structure of pyruvate kinase (PYK) from Trypanosoma cruzi | | Descriptor: | GLYCEROL, POTASSIUM ION, Pyruvate kinase | | Authors: | Zhong, W, Morgan, H.P, McNae, I.W, Michels, P.A.M, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2013-05-17 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of pyruvate kinases display evolutionarily divergent allosteric strategies.
R Soc Open Sci, 1, 2014
|
|
4KCW
 
 | | Pyruvate kinase (PYK) from Trypanosoma brucei soaked with oxalate | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Zhong, W, Morgan, H.P, McNae, I.W, Michels, P.A.M, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2013-04-24 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Pyruvate kinases have an intrinsic and conserved decarboxylase activity.
Biochem.J., 458, 2014
|
|
4KS0
 
 | | Pyruvate kinase (PYK) from Trypanosoma cruzi in the presence of Magnesium, oxalate and F26BP | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, OXALATE ION, ... | | Authors: | Morgan, H.P, Zhong, W, McNae, I.W, Michels, P.A.M, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2013-05-17 | | Release date: | 2014-11-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of pyruvate kinases display evolutionarily divergent allosteric strategies.
R Soc Open Sci, 1, 2014
|
|
4KCV
 
 | | Pyruvate kinase (PYK) from Trypanosoma brucei soaked with 2-oxoglutaric acid | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, 2-OXOGLUTARIC ACID, GLYCEROL, ... | | Authors: | Zhong, W, Morgan, H.P, McNae, I.W, Michels, P.A.M, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2013-04-24 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Pyruvate kinases have an intrinsic and conserved decarboxylase activity.
Biochem.J., 458, 2014
|
|
5AER
 
 | | Neuronal calcium sensor-1 (NCS-1)from Rattus norvegicus complex with D2 dopamine receptor peptide from Homo sapiens | | Descriptor: | CALCIUM ION, D(2) DOPAMINE RECEPTOR, NEURONAL CALCIUM SENSOR 1, ... | | Authors: | Saleem, M, Karuppiah, V, Pandalaneni, S, Burgoyne, R, Derrick, J.P, Lian, L.Y. | | Deposit date: | 2015-01-08 | | Release date: | 2015-02-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Neuronal Calcium Sensor-1 Binds the D2 Dopamine Receptor and G-Protein-Coupled Receptor Kinase 1 (Grk1) Peptides Using Different Modes of Interactions.
J.Biol.Chem., 290, 2015
|
|
