6TQI
 
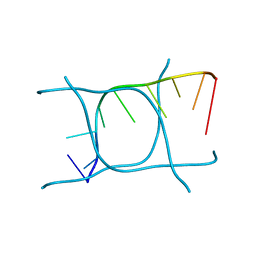 | | I-MOTIF STRUCTURE FORMED FROM THE C STRAND OF A HUMAN TELOMERE FRAGMENT | | Descriptor: | DNA (5'-*TP*AP*AP*CP*CP*CP*TP*AP*A-3') | | Authors: | Parkinson, G.N, Wagner, A, Viladoms-Claverol, J, Duman, R, El-Omari, K. | | Deposit date: | 2019-12-16 | | Release date: | 2020-06-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Native de novo structural determinations of non-canonical nucleic acid motifs by X-ray crystallography at long wavelengths.
Nucleic Acids Res., 48, 2020
|
|
4UBL
 
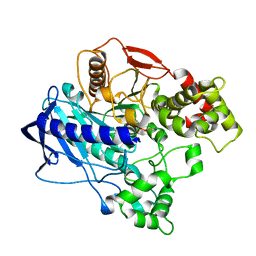 | | KINETIC CRYSTALLOGRAPHY OF ALPHA_E7-CARBOXYLESTERSE FROM LUCILLA CUPRINA - ABSORBED X-RAY DOSE 9.26 MGy | | Descriptor: | DIETHYL HYDROGEN PHOSPHATE, E3 | | Authors: | Jackson, C.J, Carr, P.D, Weik, M, Huber, T, Meirelles, T, Correy, G. | | Deposit date: | 2014-08-13 | | Release date: | 2015-08-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Mapping the Accessible Conformational Landscape of an Insect Carboxylesterase Using Conformational Ensemble Analysis and Kinetic Crystallography
Structure, 24, 2016
|
|
8AHL
 
 | |
4UBW
 
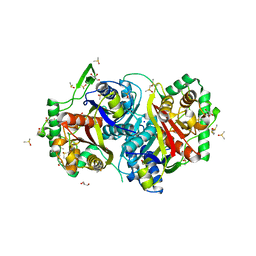 | |
4UDG
 
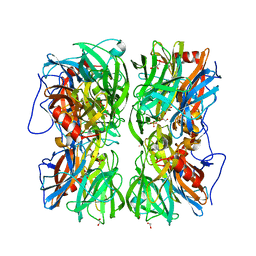 | | Crystal structure of b-1,4-mannopyranosyl-chitobiose phosphorylase at 1.60 Angstrom in complex with N-acetylglucosamine and inorganic phosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose, GLYCEROL, ... | | Authors: | Ladeveze, S, Cioci, G, Potocki-Veronese, G, Tranier, S, Mourey, L. | | Deposit date: | 2014-12-10 | | Release date: | 2015-05-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Bases for N-Glycan Processing by Mannoside Phosphorylase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UMF
 
 | | Crystal structure of 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase from Moraxella catarrhalis in complex with Magnesium ion, Phosphate ion and KDO molecule | | Descriptor: | 3-DEOXY-D-MANNO-OCTULOSONATE 8-PHOSPHATE PHOSPHATASE KDSC, 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, MAGNESIUM ION, ... | | Authors: | Dhindwal, S, Tomar, S, Kumar, P. | | Deposit date: | 2014-05-16 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Ligand-Bound Structures of 3-Deoxy-D-Manno-Octulosonate 8-Phosphate Phosphatase from Moraxella Catarrhalis Reveal a Water Channel Connecting to the Active Site for the Second Step of Catalysis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UNR
 
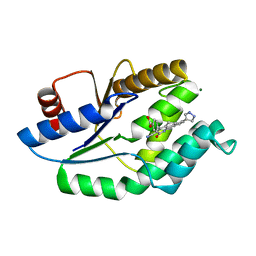 | | Mtb TMK in complex with compound 23 | | Descriptor: | 4-[3-cyano-2-oxo-7-(1H-pyrazol-4-yl)-5,6-dihydro-1H-benzo[h]quinolin-4-yl]benzoic acid, MAGNESIUM ION, Thymidylate kinase | | Authors: | Read, J.A, Hussein, S, Gingell, H, Tucker, J. | | Deposit date: | 2014-05-30 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure Guided Lead Generation for M. Tuberculosis Thymidylate Kinase (Mtb Tmk): Discovery of 3-Cyanopyridone and 1,6-Naphthyridin-2-One as Potent Inhibitors.
J.Med.Chem., 58, 2015
|
|
4UNY
 
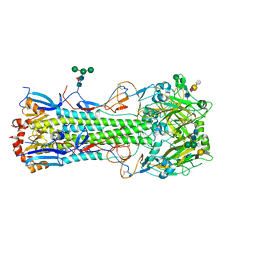 | | Structure of the A_Equine_Newmarket_2_93 H3 haemagglutinin in complex with 6SO4-3SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, H3 HAEMAGGLUTININ HA2 CHAIN, ... | | Authors: | Vachieri, S.G, Collins, P.J, Haire, L.F, Ogrodowicz, R.W, Martin, S.R, Walker, P.A, Xiong, X, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2014-05-31 | | Release date: | 2014-07-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Recent Evolution of Equine Influenza and the Origin of Canine Influenza.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8AFH
 
 | |
4UN3
 
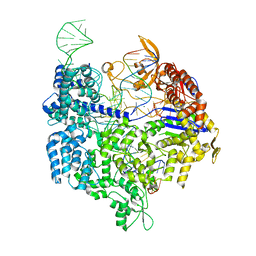 | | Crystal structure of Cas9 bound to PAM-containing DNA target | | Descriptor: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MAGNESIUM ION, NON-TARGET DNA STRAND, ... | | Authors: | Anders, C, Niewoehner, O, Duerst, A, Jinek, M. | | Deposit date: | 2014-05-25 | | Release date: | 2014-07-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.593 Å) | | Cite: | Structural Basis of Pam-Dependent Target DNA Recognition by the Cas9 Endonuclease
Nature, 513, 2014
|
|
4UQK
 
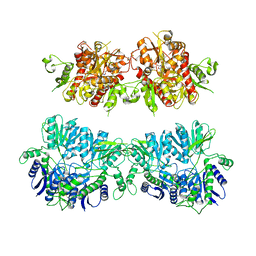 | | Electron density map of GluA2em in complex with quisqualate and LY451646 | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, GLUTAMATE RECEPTOR 2 | | Authors: | Meyerson, J.R, Kumar, J, Chittori, S, Rao, P, Pierson, J, Bartesaghi, A, Mayer, M.L, Subramaniam, S. | | Deposit date: | 2014-06-24 | | Release date: | 2014-08-13 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (16.4 Å) | | Cite: | Structural Mechanism of Glutamate Receptor Activation and Desensitization
Nature, 514, 2014
|
|
6TEK
 
 | | Structure of siderophore interaction domain of IrtAB | | Descriptor: | Drug ABC transporter ATP-binding protein, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Arnold, F.M, Gonda, I, Hutter, C.A.J, Seeger, M.A, Hurlimann, L.M. | | Deposit date: | 2019-11-12 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The ABC exporter IrtAB imports and reduces mycobacterial siderophores.
Nature, 580, 2020
|
|
4UER
 
 | | 40S-eIF1-eIF1A-eIF3-eIF3j translation initiation complex from Lachancea kluyveri | | Descriptor: | 18S RRNA, EIF1, EIF1A, ... | | Authors: | Aylett, C.H.S, Boehringer, D, Erzberger, J.P, Schaefer, T, Ban, N. | | Deposit date: | 2014-12-18 | | Release date: | 2015-02-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.47 Å) | | Cite: | Structure of a Yeast 40S-Eif1-Eif1A-Eif3-Eif3J Initiation Complex
Nat.Struct.Mol.Biol., 22, 2015
|
|
4UB9
 
 | | Structural and catalytic characterization of molinate hydrolase | | Descriptor: | Molinate hydrolase, ZINC ION | | Authors: | Leite, J.P, Duarte, M, Paiva, A, Ferreira-da-Silva, F, Matias, P.M, Nunes, O, Gales, L. | | Deposit date: | 2014-08-12 | | Release date: | 2015-06-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure-guided engineering of molinate hydrolase for the degradation of thiocarbamate pesticides.
Plos One, 10, 2015
|
|
4UI8
 
 | | Crystal structure of human tankyrase 2 in complex with TA-55 | | Descriptor: | 8-HYDROXY-2-[4-(TRIFLUOROMETHYL)PHENYL]-3,4-DIHYDROQUINAZOLIN-4-ONE, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Lehtio, L. | | Deposit date: | 2015-03-27 | | Release date: | 2016-04-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-Activity Relationships of 2-Arylquinazolin-4-Ones as Highly Selective and Potent Inhibitors of the Tankyrases.
Eur.J.Med.Chem., 118, 2016
|
|
4UHI
 
 | | HUMAN STEROL 14-ALPHA DEMETHYLASE (CYP51) IN COMPLEX WITH VFV IN C121 SPACE GROUP | | Descriptor: | N-[(1R)-1-(3,4'-difluorobiphenyl-4-yl)-2-(1H-imidazol-1-yl)ethyl]-4-(5-phenyl-1,3,4-oxadiazol-2-yl)benzamide, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE | | Authors: | Hargrove, T.Y, Wawrzak, Z, I Lepesheva, G. | | Deposit date: | 2015-03-24 | | Release date: | 2016-06-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Human Sterol 14Alpha-Demethylase (Cyp51) as a Target for Anticancer Chemotherapy: Towards Structure-Aided Drug Design.
J.Lipid Res., 57, 2016
|
|
6F9Y
 
 | |
4UCT
 
 | | Fragment bound to H.influenza NAD dependent DNA ligase | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 2-amino-6-methyl-5-(propan-2-yloxy)-3H-[1,2,4]triazolo[1,5-a]pyrimidin-8-ium, DNA LIGASE | | Authors: | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
4UCX
 
 | | Structure of the T18G small subunit mutant of D. fructosovorans NiFe- hydrogenase | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Abou-Hamdan, A, Ceccaldi, P, Lebrette, H, Guttierez-Sanz, O, Richaud, P, Cournac, L, Guigliarelli, B, deLacey, A.L, Leger, C, Volbeda, A, Burlat, B, Dementin, S. | | Deposit date: | 2014-12-05 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A threonine stabilizes the NiC and NiR catalytic intermediates of [NiFe]-hydrogenase.
J. Biol. Chem., 290, 2015
|
|
4UDV
 
 | | Cryo-EM structure of TMV at 3.35 A resolution | | Descriptor: | 5'-D(*GP*AP*AP)-3', CAPSID PROTEIN | | Authors: | Fromm, S.A, Bharat, T.A.M, Jakobi, A.J, Hagen, W.J.H, Sachse, C. | | Deposit date: | 2014-12-11 | | Release date: | 2014-12-31 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Seeing Tobacco Mosaic Virus Through Direct Electron Detectors.
J.Struct.Biol., 189, 2015
|
|
4UGM
 
 | | Structure of Bacillus subtilis Nitric Oxide Synthase in complex with N,N'-(ethane-1,2-diyldibenzene-3,1-diyl)dithiophene-2-carboximidamide | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, CHLORIDE ION, N,N'-(ethane-1,2-diyldibenzene-3,1-diyl)dithiophene-2-carboximidamide, ... | | Authors: | Holden, J.K, Poulos, T.L. | | Deposit date: | 2015-03-22 | | Release date: | 2015-06-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Inhibitor Bound Crystal Structures of Bacterial Nitric Oxide Synthase.
Biochemistry, 54, 2015
|
|
4UI7
 
 | | Crystal structure of human tankyrase 2 in complex with TA-49 | | Descriptor: | 8-HYDROXY-2-(4-METHYLPHENYL)-3,4-DIHYDROQUINAZOLIN-4-ONE, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Lehtio, L. | | Deposit date: | 2015-03-27 | | Release date: | 2016-04-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Activity Relationships of 2-Arylquinazolin-4-Ones as Highly Selective and Potent Inhibitors of the Tankyrases.
Eur.J.Med.Chem., 118, 2016
|
|
4ULL
 
 | | SOLUTION NMR STRUCTURE OF VEROTOXIN-1 B-SUBUNIT FROM E. COLI, 5 STRUCTURES | | Descriptor: | Shiga toxin 1B | | Authors: | Richardson, J.M, Evans, P.D, Homans, S.W, Donohue-Rolfe, A. | | Deposit date: | 1996-12-17 | | Release date: | 1997-04-01 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the carbohydrate-binding B-subunit homopentamer of verotoxin VT-1 from E. coli.
Nat.Struct.Biol., 4, 1997
|
|
6Z6A
 
 | | Keap1 macrocycle complex | | Descriptor: | (5S,8R)-8-[[(2S)-1-ethanoylpyrrolidin-2-yl]carbonylamino]-N,N-dimethyl-7,11-bis(oxidanylidene)-10-oxa-3-thia-6-azabicyclo[10.4.0]hexadeca-1(16),12,14-triene-5-carboxamide, CHLORIDE ION, Kelch-like ECH-associated protein 1, ... | | Authors: | Johansson, P. | | Deposit date: | 2020-05-28 | | Release date: | 2020-12-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Mining Natural Products for Macrocycles to Drug Difficult Targets.
J.Med.Chem., 64, 2021
|
|
4UM3
 
 | | Engineered Ls-AChBP with alpha4-alpha4 binding pocket in complex with NS3920 | | Descriptor: | 1-(6-bromopyridin-3-yl)-1,4-diazepane, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINE BINDING PROTEIN, ... | | Authors: | Shahsavar, A, Kastrup, J.S, Balle, T, Gajhede, M. | | Deposit date: | 2014-05-14 | | Release date: | 2015-07-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Achbp Engineered to Mimic the Alpha4-Alpha4 Binding Pocket in Alpha4Beta2 Nicotinic Acetylcholine Receptors Reveals Interface Specific Interactions Important for Binding and Activity
Mol.Pharmacol., 88, 2015
|
|
