4M0N
 
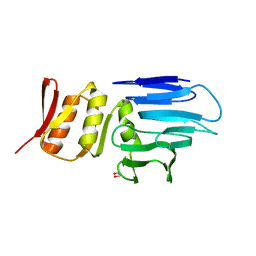 | |
4DRT
 
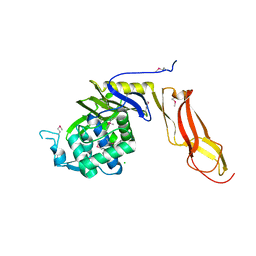 | | Three dimensional structure of de novo designed serine hydrolase OSH26, Northeast Structural Genomics Consortium (NESG) target OR89 | | Descriptor: | CHLORIDE ION, SODIUM ION, de novo designed serine hydrolase, ... | | Authors: | Kuzin, A, Su, M, Rajagopalan, S, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-02-17 | | Release date: | 2012-04-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
4LZW
 
 | | X-ray structure uridine phosphorylase from Vibrio cholerae in complex with thymidine at 1.29 A resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, ETHANOL, ... | | Authors: | Prokofev, I.I, Lashkov, A.A, Gabdoulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2013-08-01 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | X-ray structures of uridine phosphorylase from Vibrio cholerae in complexes with uridine, thymidine, uracil, thymine, and phosphate anion: Substrate specificity of bacterial uridine phosphorylases
Crystallography Reports, 2016
|
|
4DS4
 
 | | Ternary complex of Bacillus DNA Polymerase I Large Fragment, DNA duplex, and rCTP in presence of Mn2+ | | Descriptor: | DNA (5'-D(*CP*AP*TP*GP*GP*GP*AP*GP*TP*CP*AP*GP*G)-3'), DNA (5'-D(*CP*CP*TP*GP*AP*CP*TP*CP*(DOC))-3'), DNA polymerase, ... | | Authors: | Wang, W, Beese, L.S. | | Deposit date: | 2012-02-17 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.681 Å) | | Cite: | Structural factors that determine selectivity of a high fidelity DNA polymerase for deoxy-, dideoxy-, and ribonucleotides.
J.Biol.Chem., 287, 2012
|
|
4M1J
 
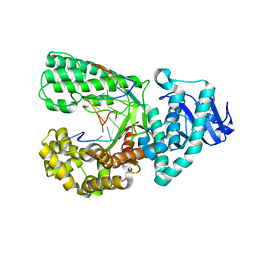
 | | Crystal structure of Pseudomonas aeruginosa PvdQ in complex with a transition state analogue | | Descriptor: | Acyl-homoserine lactone acylase PvdQ subunit alpha, Acyl-homoserine lactone acylase PvdQ subunit beta, GLYCEROL, ... | | Authors: | Wu, R, Clevenger, K, Er, J, Fast, W.L, Liu, D. | | Deposit date: | 2013-08-02 | | Release date: | 2013-08-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rational Design of a Transition State Analogue with Picomolar Affinity for Pseudomonas aeruginosa PvdQ, a Siderophore Biosynthetic Enzyme.
Acs Chem.Biol., 8, 2013
|
|
4DNH
 
 | | Crystal structure of hypothetical protein SMc04132 from Sinorhizobium meliloti 1021 | | Descriptor: | GLYCEROL, Uncharacterized protein | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-02-08 | | Release date: | 2012-03-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of hypothetical protein SMc04132 from Sinorhizobium meliloti 1021
To be Published
|
|
4M1S
 
 | | Crystal Structure of small molecule vinylsulfonamide 13 covalently bound to K-Ras G12C | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, K-Ras GTPase, N-{1-[N-(2,4-dichlorophenyl)glycyl]piperidin-4-yl}ethanesulfonamide | | Authors: | Ostrem, J.M, Peters, U, Sos, M.L, Wells, J.A, Shokat, K.M. | | Deposit date: | 2013-08-04 | | Release date: | 2013-11-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions.
Nature, 503, 2013
|
|
4M47
 
 | | structure of human DNA polymerase complexed with 8-BrG in the template base paired with incoming non-hydrolyzable GTP | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA polymerase beta, MAGNESIUM ION, ... | | Authors: | Koag, M.C, Min, K, Monzingo, A.F, Lee, S. | | Deposit date: | 2013-08-06 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural basis for promutagenicity of 8-halogenated Guanine.
J.Biol.Chem., 289, 2014
|
|
4DON
 
 | | Brd4 Bromodomain 1 complex with a fragment 3,4-Dihydro-3-methyl-2(1H)-quinazolinon | | Descriptor: | 3-methyl-3,4-dihydroquinazolin-2(1H)-one, Bromodomain-containing protein 4 | | Authors: | Xiong, B, Cao, D.Y, Chen, W.Y, Chen, T.T, Xu, Y.C, Shen, J.K. | | Deposit date: | 2012-02-09 | | Release date: | 2013-02-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Brd4 Bromodomain 1 complex with a fragment 3,4-Dihydro-3-methyl-2(1H)-quinazolinon
To be Published
|
|
4M4X
 
 | |
4DPW
 
 | | Crystal structure of Staphylococcus epidermidis D283A mevalonate diphosphate decarboxylase complexed with mevalonate diphosphate and ATPgS | | Descriptor: | (3R)-3-HYDROXY-5-{[(R)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}-3-METHYLPENTANOIC ACID, FORMIC ACID, GLYCEROL, ... | | Authors: | Barta, M.L, McWhorter, W.J, Geisbrecht, B.V. | | Deposit date: | 2012-02-14 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | Structural basis for nucleotide binding and reaction catalysis in mevalonate diphosphate decarboxylase.
Biochemistry, 51, 2012
|
|
4DQV
 
 | |
4DR5
 
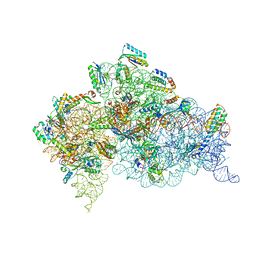 | | Crystal structure of the Thermus thermophilus (HB8) 30S ribosomal subunit with codon, crystallographically disordered cognate transfer RNA anticodon stem-loop and streptomycin bound | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2012-02-16 | | Release date: | 2012-11-14 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | A structural basis for streptomycin-induced misreading of the genetic code.
Nat Commun, 4, 2013
|
|
4DWB
 
 | | Crystal structure of Trypanosoma cruzi farnesyl diphosphate synthase in complex with [2-(n-pentylamino)ethane-1,1-diyl]bisphosphonic acid and Mg2+ | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, ACETATE ION, Farnesyl pyrophosphate synthase, ... | | Authors: | Aripirala, S, Amzel, L.M, Gabelli, S.B. | | Deposit date: | 2012-02-24 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, synthesis, calorimetry, and crystallographic analysis of 2-alkylaminoethyl-1,1-bisphosphonates as inhibitors of Trypanosoma cruzi farnesyl diphosphate synthase.
J.Med.Chem., 55, 2012
|
|
4DA9
 
 | | Crystal structure of putative Short-chain dehydrogenase/reductase from Sinorhizobium meliloti 1021 | | Descriptor: | SULFATE ION, Short-chain dehydrogenase/reductase | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-01-12 | | Release date: | 2012-01-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of putative Short-chain dehydrogenase/reductase from Sinorhizobium meliloti 1021
To be Published
|
|
4MDL
 
 | | Meta Carborane Carbonic Anhydrase Inhibitor | | Descriptor: | 1-(sulfamoylamino)methyl-1,7-dicarba-closo-dodecaborane, Carbonic anhydrase 2, MERCURIBENZOIC ACID, ... | | Authors: | Brynda, J, Mader, P, Rezacova, P, Cigler, P. | | Deposit date: | 2013-08-23 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.516 Å) | | Cite: | Carborane-based carbonic anhydrase inhibitors.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
4DDJ
 
 | |
4DE2
 
 | | CTX-M-9 class A beta-lactamase complexed with compound 12 | | Descriptor: | 3-[(dimethylamino)methyl]-N-[3-(1H-tetrazol-5-yl)phenyl]benzamide, Beta-lactamase, DIMETHYL SULFOXIDE | | Authors: | Nichols, D.A, Chen, Y. | | Deposit date: | 2012-01-19 | | Release date: | 2012-03-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Based Design of Potent and Ligand-Efficient Inhibitors of CTX-M Class A Beta-Lactamase
J.Med.Chem., 55, 2012
|
|
4MFR
 
 | | Crystal structure of Mycobacterium tuberculosis CarD | | Descriptor: | GLYCEROL, IODIDE ION, RNA polymerase-binding transcription factor CarD, ... | | Authors: | Kaur, G, Thakur, K.G. | | Deposit date: | 2013-08-28 | | Release date: | 2013-11-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis CarD, an essential RNA polymerase binding protein, reveals a quasidomain-swapped dimeric structural architecture.
Proteins, 82, 2014
|
|
4LX5
 
 | | X-ray crystal structure of the M6" riboswitch aptamer bound to pyrimido[4,5-d]pyrimidine-2,4-diamine (PPDA) | | Descriptor: | MAGNESIUM ION, Mutated adenine riboswitch aptamer, pyrimido[4,5-d]pyrimidine-2,4-diamine | | Authors: | Dunstan, M.S, Leys, D. | | Deposit date: | 2013-07-29 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Modular riboswitch toolsets for synthetic genetic control in diverse bacterial species.
J.Am.Chem.Soc., 136, 2014
|
|
4DGC
 
 | | TRIMCyp cyclophilin domain from Macaca mulatta: cyclosporin A complex | | Descriptor: | TRIMCyp, cyclosporin A | | Authors: | Caines, M.E.C, Bichel, K, Price, A.J, McEwan, W.A, James, L.C. | | Deposit date: | 2012-01-25 | | Release date: | 2012-02-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Diverse HIV viruses are targeted by a conformationally dynamic antiviral.
Nat.Struct.Mol.Biol., 19, 2012
|
|
6VMZ
 
 | | Crystal Structure of a H5N1 influenza virus hemagglutinin with CBS1117 | | Descriptor: | 2,6-dichloro-N-[1-(propan-2-yl)piperidin-4-yl]benzamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Antanasijevic, A, Durst, M.A, Lavie, A, Caffrey, M. | | Deposit date: | 2020-01-28 | | Release date: | 2020-07-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of avian influenza hemagglutinin in complex with a small molecule entry inhibitor.
Life Sci Alliance, 3, 2020
|
|
4DGT
 
 | | Crystal structure of PLP-bound putative aminotransferase from Clostridium difficile 630 crystallized with magnesium formate | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Shabalin, I.G, Onopriyenko, O, Kudritska, M, Chruszcz, M, Grimshaw, S, Porebski, P.J, Cooper, D.R, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-01-26 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of putative aminotransferase from Clostridium difficile
630
to be published
|
|
4DJ1
 
 | |
4DJN
 
 | |
