6NR5
 
 | | Human LSD1 in complex with Phenelzine sulfate | | Descriptor: | Lysine-specific histone demethylase 1A, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R,3R,4R)-5-[(4aR)-7,8-dimethyl-2,4-dioxo-5-(2-phenylethyl)-3,4,4a,5-tetrahydrobenzo[g]pteridin-10(2H)-yl]-2,3,4-trihydroxypentyl dihydrogen diphosphate (non-preferred name) | | Authors: | Tan, A.H.Y, Tu, W, McCuaig, R, Donovan, T, Tsimbalyuk, S, Forwood, J.K, Rao, S. | | Deposit date: | 2019-01-22 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Lysine-Specific Histone Demethylase 1A Regulates Macrophage Polarization and Checkpoint Molecules in the Tumor Microenvironment of Triple-Negative Breast Cancer.
Front Immunol, 10, 2019
|
|
8VK9
 
 | | Structure of UbV.d2.3 in complex with Ube2d2-S22R | | Descriptor: | GLYCEROL, Ubiquitin variant D2.3, Ubiquitin-conjugating enzyme E2 D2 | | Authors: | Middleton, A.J. | | Deposit date: | 2024-01-08 | | Release date: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and biophysical characterisation of ubiquitin variants that inhibit the ubiquitin conjugating enzyme Ube2d2
To Be Published
|
|
7V0G
 
 | | Structure of cAMP-dependent protein kinase using a MD-MX procedure, produced using 1.63 Angstrom data | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Wych, D.C, Aoto, P.C, Wall, M.E. | | Deposit date: | 2022-05-10 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Molecular-dynamics simulation methods for macromolecular crystallography.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8Q3H
 
 | | Capra hircus reactive intermediate deaminase A mutant - A108D | | Descriptor: | 1,2-ETHANEDIOL, 2-iminobutanoate/2-iminopropanoate deaminase, GLYCEROL | | Authors: | Rizzi, G, Visentin, C, Di Pisa, F, Ricagno, S. | | Deposit date: | 2023-08-04 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Capra hircus reactive intermediate deaminase A mutant - A108D
To Be Published
|
|
6O0G
 
 | | M.tb MenD bound to Intermediate I and Inhibitor | | Descriptor: | 1,4-dihydroxy-2-naphthoic acid, 2-OXOGLUTARIC ACID, 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase, ... | | Authors: | Johnston, J.M, Bashiri, G, Bulloch, E.M.M, Jirgis, E.M.N, Chuang, H, Nigon, L.V, Baker, E.N. | | Deposit date: | 2019-02-16 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Allosteric regulation of menaquinone (vitamin K2) biosynthesis in the human pathogenMycobacterium tuberculosis.
J.Biol.Chem., 295, 2020
|
|
8WDM
 
 | | Crystal structure of a novel PU plastic degradation enzyme from Thermaerobacter marianensis | | Descriptor: | Carboxylic ester hydrolase | | Authors: | Li, Z.S, Wang, H, Gao, J, Chen, Y.Y, Wei, H.L, Li, Q, Han, X, Wei, R, Liu, W.D. | | Deposit date: | 2023-09-15 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a novel PU plastic degradation enzyme from Thermaerobacter marianensis
To Be Published
|
|
7OBF
 
 | |
7UYG
 
 | |
8WC1
 
 | |
8QH1
 
 | |
6NRP
 
 | | Putative short-chain dehydrogenase/reductase (SDR) from Acinetobacter baumannii | | Descriptor: | 3-oxoacyl-ACP reductase FabG | | Authors: | Cross, E.M, Smith, K.M, Shaw, K.I, Aragao, D, Forwood, J.K. | | Deposit date: | 2019-01-23 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into Acinetobacter baumannii fatty acid synthesis 3-oxoacyl-ACP reductases.
Sci Rep, 11, 2021
|
|
5E97
 
 | | Glycoside Hydrolase ligand structure 1 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Wu, L, Davies, G.J. | | Deposit date: | 2015-10-14 | | Release date: | 2015-11-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural characterization of human heparanase reveals insights into substrate recognition.
Nat.Struct.Mol.Biol., 22, 2015
|
|
7ZRP
 
 | | 2.65 Angstrom crystal structure of Ca/CaM:CaMKIIdelta peptide complex | | Descriptor: | CALCIUM ION, Calcium/calmodulin-dependent protein kinase type II subunit delta, Calmodulin-1, ... | | Authors: | Helassa, N, Antonyuk, S. | | Deposit date: | 2022-05-04 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Calmodulin variant E140G associated with long QT syndrome impairs CaMKII delta autophosphorylation and L-type calcium channel inactivation.
J.Biol.Chem., 299, 2022
|
|
5E9I
 
 | |
6O0W
 
 | | Crystal structure of the TIR domain from the grapevine disease resistance protein RUN1 in complex with NADP+ and Bis-Tris | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-2'-5'-DIPHOSPHATE, TIR-NB-LRR type resistance protein RUN1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
8QOQ
 
 | |
7Q4T
 
 | |
6NSI
 
 | | Crystal structure of Fe(III)-bound YtgA from Chlamydia trachomatis | | Descriptor: | CALCIUM ION, FE (III) ION, Manganese-binding protein, ... | | Authors: | Luo, Z, Campbell, R, Begg, S.L, Kobe, B, McDevitt, C.A. | | Deposit date: | 2019-01-24 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.00006342 Å) | | Cite: | Structure and Metal Binding Properties of Chlamydia trachomatis YtgA.
J.Bacteriol., 202, 2019
|
|
7ZRQ
 
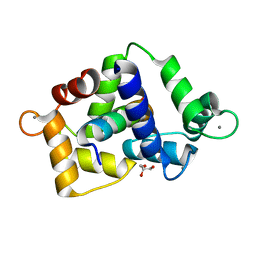 | | 1.68 Angstrom crystal structure of Ca/CaM-E140G:CaMKIIdelta peptide complex | | Descriptor: | CALCIUM ION, Calcium/calmodulin-dependent protein kinase type II subunit delta, Calmodulin-1, ... | | Authors: | Helassa, N, Antonyuk, S. | | Deposit date: | 2022-05-04 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Calmodulin variant E140G associated with long QT syndrome impairs CaMKII delta autophosphorylation and L-type calcium channel inactivation.
J.Biol.Chem., 299, 2022
|
|
7QGA
 
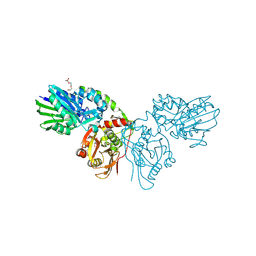 | |
7U6L
 
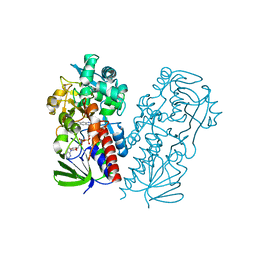 | | Pseudooxynicotine amine oxidase | | Descriptor: | Amine oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Choudhary, V, Stull, F, Wu, K. | | Deposit date: | 2022-03-04 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The enzyme pseudooxynicotine amine oxidase from Pseudomonas putida S16 is not an oxidase, but a dehydrogenase.
J.Biol.Chem., 298, 2022
|
|
8SI0
 
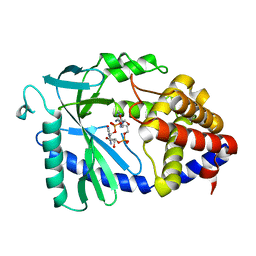 | |
7MWL
 
 | | The TAM domain of BAZ2A in complex with a 12mer mCG DNA | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, DNA (5'-D(*GP*CP*CP*AP*AP*(5CM)P*GP*TP*TP*GP*GP*C)-3'), GLYCEROL | | Authors: | Liu, K, Dong, A, Li, Y, Loppnau, P, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-17 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The TAM domain of BAZ2A in complex with a 12mer mCG DNA
To Be Published
|
|
8PZ6
 
 | | crystal structure of VDR in complex with D-Bishomo-1a,25-dihydroxyvitamin D3 analog 56 | | Descriptor: | (1~{R},3~{R})-5-[(2~{E})-2-[(4~{a}~{R},5~{S},9~{a}~{S})-4~{a}-methyl-5-[(2~{R})-6-methyl-6-oxidanyl-heptan-2-yl]-3,4,5,6,7,8,9,9~{a}-octahydro-2~{H}-benzo[7]annulen-1-ylidene]ethylidene]-2-(3-oxidanylpropylidene)cyclohexane-1,3-diol, Nuclear receptor coactivator 2, Vitamin D3 receptor A | | Authors: | Rochel, N. | | Deposit date: | 2023-07-27 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design, synthesis, and biological activity of D-bishomo-1 alpha ,25-dihydroxyvitamin D 3 analogs and their crystal structures with the vitamin D nuclear receptor.
Eur.J.Med.Chem., 271, 2024
|
|
8SHY
 
 | |
