8TVN
 
 | | IRAK4 in complex with compound 23 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, N-{1-[(1R,2S)-2-fluorocyclopropyl]-2-oxo-1,2-dihydropyridin-3-yl}-2-[(1R,4r)-1-methyl-2-oxabicyclo[2.1.1]hexan-4-yl]-6-[(propan-2-yl)oxy]-2H-pyrazolo[3,4-b]pyridine-5-carboxamide | | Authors: | Metrick, C.M, Chodaparambil, J.V. | | Deposit date: | 2023-08-18 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Tiny Pocket Packs a Punch: Leveraging Pyridones for the Discovery of CNS-Penetrant Aza-indazole IRAK4 Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
8TVM
 
 | | IRAK4 in complex with compound 24 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, N-{1-[(1R,2R)-2-fluorocyclopropyl]-2-oxo-1,2-dihydropyridin-3-yl}-2-[(1R,4r)-1-methyl-2-oxabicyclo[2.1.1]hexan-4-yl]-6-[(propan-2-yl)oxy]-2H-pyrazolo[3,4-b]pyridine-5-carboxamide | | Authors: | Metrick, C.M, Chodaparambil, J.V. | | Deposit date: | 2023-08-18 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Tiny Pocket Packs a Punch: Leveraging Pyridones for the Discovery of CNS-Penetrant Aza-indazole IRAK4 Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
6LXY
 
 | | IRAK4 in complex with inhibitor | | Descriptor: | Interleukin-1 receptor-associated kinase 4, N-[(2R)-2-fluoranyl-3-methyl-3-oxidanyl-butyl]-6-[(6-fluoranylpyrazolo[1,5-a]pyrimidin-5-yl)amino]-4-(propan-2-ylamino)pyridine-3-carboxamide, SULFATE ION | | Authors: | Ghosh, K, Bose, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-11-25 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Optimization of Nicotinamides as Potent and Selective IRAK4 Inhibitors with Efficacy in a Murine Model of Psoriasis.
Acs Med.Chem.Lett., 11, 2020
|
|
5QJ2
 
 | | CRYSTAL STRUCTURE OF MYELOPEROXIDASE SUBFORM C (MPO) OMPLEX WITH COMPOUND-20 AKA 7-((3-(1-METHYL-1H-PYRAZOL-3- YL)BENZYL)OXY)- 1H-[1,2,3]TRIAZOLO[4,5-B]PYRIDIN-5-AMINE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-{[3-(1-methyl-1H-pyrazol-3-yl)phenyl]methoxy}-1H-[1,2,3]triazolo[4,5-b]pyridin-5-amine, CALCIUM ION, ... | | Authors: | Khan, J.A. | | Deposit date: | 2018-09-26 | | Release date: | 2019-02-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Potent Triazolopyridine Myeloperoxidase Inhibitors.
ACS Med Chem Lett, 9, 2018
|
|
6US2
 
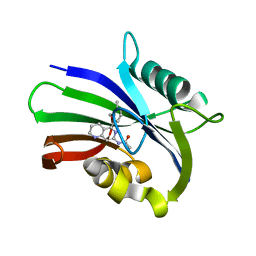 | | MTH1 in complex with compound 5 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, N-[5-(2,3-dimethylphenyl)-1,2,3,4-tetrahydro-1,6-naphthyridin-7-yl]acetamide | | Authors: | Newby, Z.E.R, Lansdon, E.B. | | Deposit date: | 2019-10-24 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.80012655 Å) | | Cite: | Discovery of Potent and Selective MTH1 Inhibitors for Oncology: Enabling Rapid Target (In)Validation.
Acs Med.Chem.Lett., 11, 2020
|
|
6US4
 
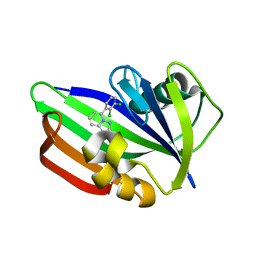 | | MTH1 in complex with compound 32 | | Descriptor: | 5-(2,3-dichlorophenyl)[1,2,4]triazolo[1,5-a]pyridin-2-amine, 7,8-dihydro-8-oxoguanine triphosphatase | | Authors: | Newby, Z.E.R, Lansdon, E.B. | | Deposit date: | 2019-10-24 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95032907 Å) | | Cite: | Discovery of Potent and Selective MTH1 Inhibitors for Oncology: Enabling Rapid Target (In)Validation.
Acs Med.Chem.Lett., 11, 2020
|
|
5QJ3
 
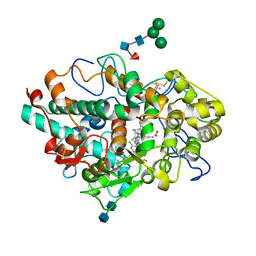 | | CRYSTAL STRUCTURE OF MYELOPEROXIDASE SUBFORM C (MPO) COMPLEX WITH COMPOUND-24 AKA 7-({4-CHLORO-3'-FLUORO-[1,1'- BIPHENYL]-3-YL}METHOXY)-3H-[1,2,3]TRIAZOLO[4,5-B]PYRIDIN- 5-AMINE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-[(4-chloro-3'-fluoro[1,1'-biphenyl]-3-yl)methoxy]-1H-[1,2,3]triazolo[4,5-b]pyridin-5-amine, ... | | Authors: | Khan, J.A. | | Deposit date: | 2018-09-26 | | Release date: | 2019-02-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Potent Triazolopyridine Myeloperoxidase Inhibitors.
ACS Med Chem Lett, 9, 2018
|
|
6US3
 
 | | MTH1 in complex with compound 4 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, N-[5-(2,3-dimethylphenyl)-1,6-naphthyridin-7-yl]acetamide | | Authors: | Newby, Z.E.R, Lansdon, E.B. | | Deposit date: | 2019-10-24 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.47028923 Å) | | Cite: | Discovery of Potent and Selective MTH1 Inhibitors for Oncology: Enabling Rapid Target (In)Validation.
Acs Med.Chem.Lett., 11, 2020
|
|
6XMK
 
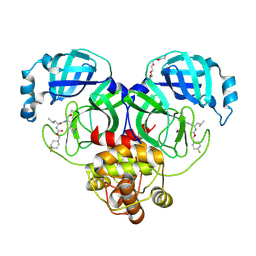 | | 1.70 A resolution structure of SARS-CoV-2 3CL protease in complex with inhibitor 7j | | Descriptor: | (1S,2S)-2-[(N-{[(4,4-difluorocyclohexyl)methoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, TETRAETHYLENE GLYCOL | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-06-30 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
8UN5
 
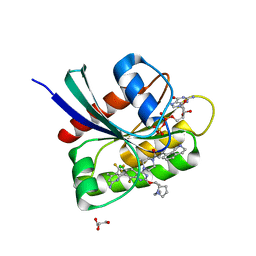 | | KRAS-G13D-GDP in complex with Cpd38 ((E)-1-((3S)-4-(7-(6-amino-4-methyl-3-(trifluoromethyl)pyridin-2-yl)-6-chloro-8-fluoro-2-(((S)-2-methylenetetrahydro-1H-pyrrolizin-7a(5H)-yl)methoxy)quinazolin-4-yl)-3-methylpiperazin-1-yl)-3-(1,2,3,4-tetrahydroisoquinolin-8-yl)prop-2-en-1-one) | | Descriptor: | (2E)-1-{(3S)-4-[(7M)-7-[6-amino-4-methyl-3-(trifluoromethyl)pyridin-2-yl]-6-chloro-8-fluoro-2-{[(4R,7aS)-2-methylidenetetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}quinazolin-4-yl]-3-methylpiperazin-1-yl}-3-(1,2,3,4-tetrahydroisoquinolin-8-yl)prop-2-en-1-one, GLYCEROL, GTPase KRas, ... | | Authors: | Ultsch, M.H. | | Deposit date: | 2023-10-18 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structure-Based Design and Evaluation of Reversible KRAS G13D Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
7SG4
 
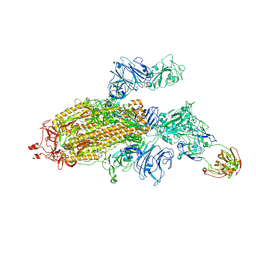 | |
6I82
 
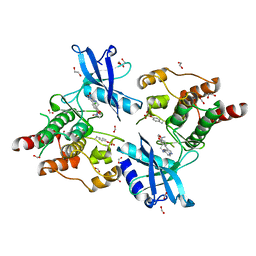 | | Crystal structure of partially phosphorylated RET V804M tyrosine kinase domain complexed with PDD00018412 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Burschowsky, D, Seewooruthun, C, Bayliss, R, Carr, M.D, Echalier, A, Jordan, A.M. | | Deposit date: | 2018-11-19 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery and Optimization of wt-RET/KDR-Selective Inhibitors of RETV804MKinase.
Acs Med.Chem.Lett., 11, 2020
|
|
8W3X
 
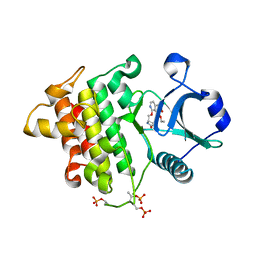 | | Crystal structure of IRAK4 in complex with compound 6 | | Descriptor: | 7-ethoxy-1-{[(2S)-5-oxopyrrolidin-2-yl]methoxy}isoquinoline-6-carboxamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Han, S, Knafels, J.D. | | Deposit date: | 2024-02-22 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.765 Å) | | Cite: | In Retrospect: Root-Cause Analysis of Structure-Activity Relationships in IRAK4 Inhibitor Zimlovisertib (PF-06650833).
Acs Med.Chem.Lett., 15, 2024
|
|
8W3W
 
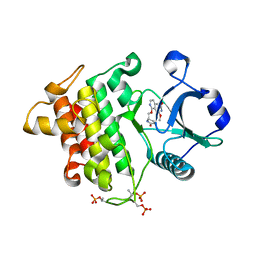 | | Crystal structure of IRAK4 in complex with compound 4 | | Descriptor: | 7-methoxy-1-{[(2S)-5-oxopyrrolidin-2-yl]methoxy}isoquinoline-6-carboxamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Han, S, Knafels, J.D. | | Deposit date: | 2024-02-22 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.976 Å) | | Cite: | In Retrospect: Root-Cause Analysis of Structure-Activity Relationships in IRAK4 Inhibitor Zimlovisertib (PF-06650833).
Acs Med.Chem.Lett., 15, 2024
|
|
4Y30
 
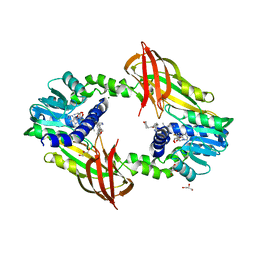 | | Crystal structure of human protein arginine methyltransferase PRMT6 bound to SAH and EPZ020411 | | Descriptor: | GLYCEROL, MAGNESIUM ION, N,N'-dimethyl-N-({3-[4-({trans-3-[2-(tetrahydro-2H-pyran-4-yl)ethoxy]cyclobutyl}oxy)phenyl]-1H-pyrazol-4-yl}methyl)ethane-1,2-diamine, ... | | Authors: | Swinger, K.K, Boriack-Sjodin, P.A. | | Deposit date: | 2015-02-10 | | Release date: | 2015-04-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Aryl Pyrazoles as Potent Inhibitors of Arginine Methyltransferases: Identification of the First PRMT6 Tool Compound.
Acs Med.Chem.Lett., 6, 2015
|
|
4UHA
 
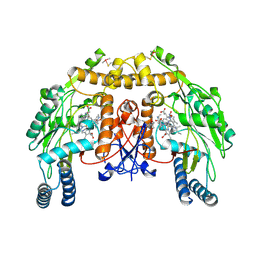 | | Structure of bovine endothelial nitric oxide synthase heme domain in complex with 3-(2-(6-Amino-4-methylpyridin-2-yl)ethyl)-5-(methyl(2-(methylamino)ethyl)amino)benzonitrile | | Descriptor: | 3-(2-(6-AMINO-4-METHYLPYRIDIN-2-YL)ETHYL)-5-(METHYL(2-(METHYLAMINO)ETHYL)AMINO)BENZONITRILE, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2015-03-23 | | Release date: | 2015-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 2-Aminopyridines with a Truncated Side Chain To Improve Human Neuronal Nitric Oxide Synthase Inhibitory Potency and Selectivity.
J. Med. Chem., 58, 2015
|
|
4Y2H
 
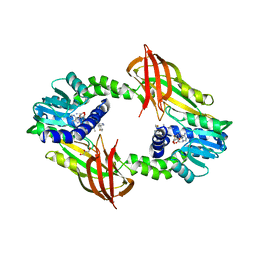 | |
6CJV
 
 | | Carbonic anhydrase IX-mimic in complex with sucralose | | Descriptor: | 4-chloro-4-deoxy-alpha-D-galactopyranose-(1-2)-1,6-dichloro-1,6-dideoxy-beta-D-fructofuranose, Carbonic anhydrase 2, ZINC ION | | Authors: | Lomelino, C.L, Murray, A.B, McKenna, R. | | Deposit date: | 2018-02-26 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.547 Å) | | Cite: | Sweet Binders: Carbonic Anhydrase IX in Complex with Sucralose.
ACS Med Chem Lett, 9, 2018
|
|
7SV8
 
 | |
7SUY
 
 | |
7SUW
 
 | |
7SV1
 
 | |
5PGZ
 
 | | CRYSTAL STRUCTURE OF MURINE 11BETA- HYDROXYSTEROIDDEHYDROGENASE COMPLEXED WITH 2-[(5R,7S)-6-HYDROXY-2-PHENYLADAMANTAN-2-YL]-1-(3-HYDROXYAZETIDIN-1-YL)ETHAN-1-ONE (BMS-816336) | | Descriptor: | 2-[(5R,7S)-6-HYDROXY-2-PHENYLADAMANTAN-2-YL]-1-(3-HYDROXYAZETIDIN-1-YL)ETHAN-1-ONE, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-02-06 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of Clinical Candidate 2-((2S,6S)-2-Phenyl-6-hydroxyadamantan-2-yl)-1-(3'-hydroxyazetidin-1-yl)ethanone [BMS-816336], an Orally Active Novel Selective 11 beta-Hydroxysteroid Dehydrogenase Type 1 Inhibitor.
J. Med. Chem., 60, 2017
|
|
5PZM
 
 | |
6I83
 
 | | Crystal structure of phosphorylated RET V804M tyrosine kinase domain complexed with PDD00018366 | | Descriptor: | 4-[5-(pyridin-3-ylmethylamino)pyrazolo[1,5-a]pyrimidin-3-yl]benzamide, FORMIC ACID, Proto-oncogene tyrosine-protein kinase receptor Ret | | Authors: | Burschowsky, D, Seewooruthun, C, Bayliss, R, Carr, M.D, Echalier, A, Jordan, A.M. | | Deposit date: | 2018-11-19 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Discovery and Optimization of wt-RET/KDR-Selective Inhibitors of RETV804MKinase.
Acs Med.Chem.Lett., 11, 2020
|
|
