3B7N
 
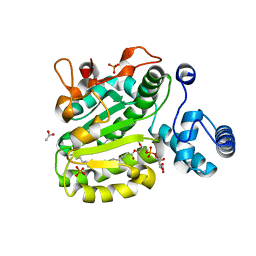 | | Crystal Structure of Yeast Sec14 Homolog Sfh1 in Complex with Phosphatidylinositol | | Descriptor: | (1R)-2-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Ortlund, E.A, Schaaf, G, Redinbo, M.R, Bankaitis, V. | | Deposit date: | 2007-10-31 | | Release date: | 2008-02-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Functional anatomy of phospholipid binding and regulation of phosphoinositide homeostasis by proteins of the sec14 superfamily
Mol.Cell, 29, 2008
|
|
3B7Z
 
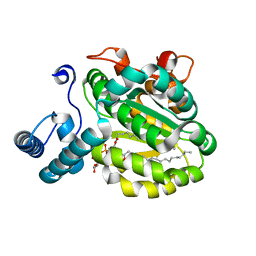 | | Crystal Structure of Yeast Sec14 Homolog Sfh1 in Complex with Phosphatidylcholine or Phosphatidylinositol | | Descriptor: | (1R)-2-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, Uncharacterized protein YKL091C | | Authors: | Ortlund, E.A, Schaaf, G, Redinbo, M.R, Bankaitis, V. | | Deposit date: | 2007-10-31 | | Release date: | 2008-02-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Functional anatomy of phospholipid binding and regulation of phosphoinositide homeostasis by proteins of the sec14 superfamily
Mol.Cell, 29, 2008
|
|
2PFI
 
 | |
3NPZ
 
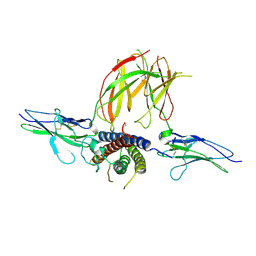 | | Prolactin Receptor (PRLR) Complexed with the Natural Hormone (PRL) | | Descriptor: | Prolactin, Prolactin receptor | | Authors: | Van Agthoven, J, England, P, Goffin, V, Broutin, I. | | Deposit date: | 2010-06-29 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structural characterization of the stem-stem dimerization interface between prolactin receptor chains complexed with the natural hormone.
J.Mol.Biol., 404, 2010
|
|
4AJ5
 
 | | Crystal structure of the Ska core complex | | Descriptor: | SPINDLE AND KINETOCHORE-ASSOCIATED PROTEIN 1, SPINDLE AND KINETOCHORE-ASSOCIATED PROTEIN 2, SPINDLE AND KINETOCHORE-ASSOCIATED PROTEIN 3 | | Authors: | Jeyaprakash, A.A, Santamaria, A, Jayachandran, U, Chan, Y.W, Benda, C, Nigg, E.A, Conti, E. | | Deposit date: | 2012-02-15 | | Release date: | 2012-05-23 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Structural and Functional Organization of the Ska Complex, a Key Component of the Kinetochore-Microtubule Interface.
Mol.Cell, 46, 2012
|
|
1SWW
 
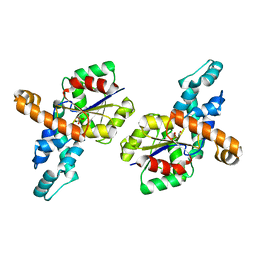 | | Crystal structure of the phosphonoacetaldehyde hydrolase D12A mutant complexed with magnesium and substrate phosphonoacetaldehyde | | Descriptor: | MAGNESIUM ION, PHOSPHONOACETALDEHYDE, phosphonoacetaldehyde hydrolase | | Authors: | Zhang, G, Morais, M.C, Dai, J, Zhang, W, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2004-03-30 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Investigation of metal ion binding in phosphonoacetaldehyde hydrolase identifies sequence markers for metal-activated enzymes of the HAD enzyme superfamily
Biochemistry, 43, 2004
|
|
7P8W
 
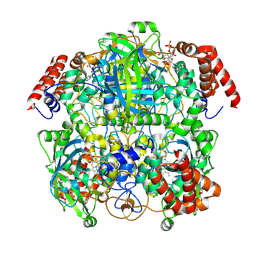 | | Human erythrocyte catalase cryoEM | | Descriptor: | Catalase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Chen, S, Li, J, Vinothkumar, K.R, Henderson, R. | | Deposit date: | 2021-07-23 | | Release date: | 2021-08-25 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Interaction of human erythrocyte catalase with air-water interface in cryoEM.
Microscopy (Oxf), 71, 2022
|
|
2G3O
 
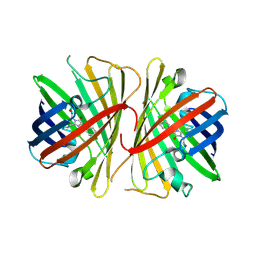 | | The 2.1A crystal structure of copGFP | | Descriptor: | green fluorescent protein 2 | | Authors: | Wilmann, P.G. | | Deposit date: | 2006-02-20 | | Release date: | 2006-08-15 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The 2.1A crystal structure of copGFP, a representative member of the copepod clade within the green fluorescent protein superfamily
J.Mol.Biol., 359, 2006
|
|
1W1I
 
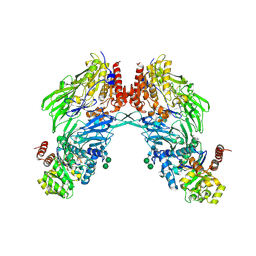 | | Crystal structure of dipeptidyl peptidase IV (DPPIV or CD26) in complex with adenosine deaminase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Weihofen, W.A, Liu, J, Reutter, W, Saenger, W, Fan, H. | | Deposit date: | 2004-06-22 | | Release date: | 2004-09-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Crystal structure of CD26/dipeptidyl-peptidase IV in complex with adenosine deaminase reveals a highly amphiphilic interface.
J. Biol. Chem., 279, 2004
|
|
7PNE
 
 | |
7PNG
 
 | |
1IAR
 
 | | INTERLEUKIN-4 / RECEPTOR ALPHA CHAIN COMPLEX | | Descriptor: | PROTEIN (INTERLEUKIN-4 RECEPTOR ALPHA CHAIN), PROTEIN (INTERLEUKIN-4) | | Authors: | Hage, T, Sebald, W, Reinemer, P. | | Deposit date: | 1999-02-25 | | Release date: | 2000-03-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the interleukin-4/receptor alpha chain complex reveals a mosaic binding interface.
Cell(Cambridge,Mass.), 97, 1999
|
|
2W86
 
 | | Crystal structure of fibrillin-1 domains cbEGF9hyb2cbEGF10, calcium saturated form | | Descriptor: | CALCIUM ION, FIBRILLIN-1, IODIDE ION | | Authors: | Jensen, S.A, Iqbal, S, Lowe, E.D, Redfield, C, Handford, P.A. | | Deposit date: | 2009-01-09 | | Release date: | 2009-05-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Interdomain Interactions of a Hybrid Domain: A Disulphide-Rich Module of the Fibrillin/Ltbp Superfamily of Matrix Proteins.
Structure, 17, 2009
|
|
1KC2
 
 | |
2OR2
 
 | | Structure of the W47A/W242A Mutant of Bacterial Phosphatidylinositol-Specific Phospholipase C | | Descriptor: | 1-phosphatidylinositol phosphodiesterase | | Authors: | Shao, C, Shi, X, Wehbi, H, Zambonelli, C, Head, J.F, Seaton, B.A, Roberts, M.F. | | Deposit date: | 2007-02-01 | | Release date: | 2007-02-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Dimer structure of an interfacially impaired phosphatidylinositol-specific phospholipase C.
J.Biol.Chem., 282, 2007
|
|
3KJR
 
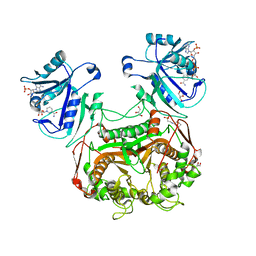 | | Crystal structure of dihydrofolate reductase/thymidylate synthase from Babesia bovis determined using SlipChip based microfluidics | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Dihydrofolate reductase/thymidylate synthase, GLYCEROL, ... | | Authors: | Li, L, Du, W, Edwards, T.E, Staker, B.L, Phan, I, Stacy, R, Ismagilov, R.F, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2009-11-03 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Multiparameter screening on SlipChip used for nanoliter protein crystallization combining free interface diffusion and microbatch methods.
J.Am.Chem.Soc., 132, 2010
|
|
1SWV
 
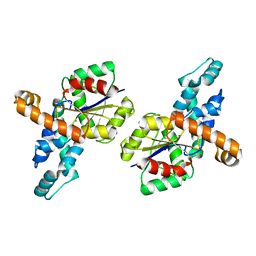 | | Crystal structure of the D12A mutant of phosphonoacetaldehyde hydrolase complexed with magnesium | | Descriptor: | MAGNESIUM ION, phosphonoacetaldehyde hydrolase | | Authors: | Zhang, G, Morais, M.C, Dai, J, Zhang, W, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2004-03-30 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Investigation of metal ion binding in phosphonoacetaldehyde hydrolase identifies sequence markers for metal-activated enzymes of the HAD enzyme superfamily
Biochemistry, 43, 2004
|
|
6MXY
 
 | |
4LTW
 
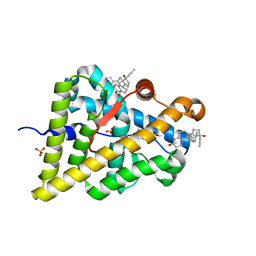 | | Ancestral Ketosteroid Receptor-Progesterone-Mifepristone Complex | | Descriptor: | 11-(4-DIMETHYLAMINO-PHENYL)-17-HYDROXY-13-METHYL-17-PROP-1-YNYL-1,2,6,7,8,11,12,13,14,15,16,17-DODEC AHYDRO-CYCLOPENTA[A]PHENANTHREN-3-ONE, Ancestral Steroid Receptor 2, GLYCEROL, ... | | Authors: | Ortlund, E.A, Colucci, J.K. | | Deposit date: | 2013-07-24 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.045 Å) | | Cite: | X-ray crystal structure of the ancestral 3-ketosteroid receptor-progesterone-mifepristone complex shows mifepristone bound at the coactivator binding interface.
Plos One, 8, 2013
|
|
6MXZ
 
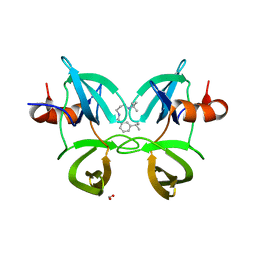 | | Structure of 53BP1 Tudor domains in complex with small molecule UNC3474 | | Descriptor: | FORMIC ACID, N-[3-(tert-butylamino)propyl]-3-(propan-2-yl)benzamide, TP53-binding protein 1 | | Authors: | Cui, G, Botuyan, M.V, Schuller, D.J, Mer, G. | | Deposit date: | 2018-10-31 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An autoinhibited state of 53BP1 revealed by small molecule antagonists and protein engineering.
Nat Commun, 14, 2023
|
|
2LCS
 
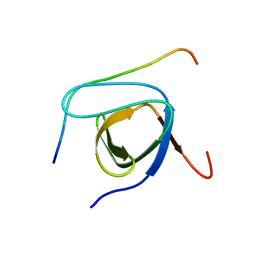 | | Yeast Nbp2p SH3 domain in complex with a peptide from Ste20p | | Descriptor: | NAP1-binding protein 2, Serine/threonine-protein kinase STE20 | | Authors: | Gorelik, M, Davidson, A.R. | | Deposit date: | 2011-05-07 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Distinct Peptide Binding Specificities of Src Homology 3 (SH3) Protein Domains Can Be Determined by Modulation of Local Energetics across the Binding Interface.
J.Biol.Chem., 287, 2012
|
|
2LXA
 
 | |
3Q7R
 
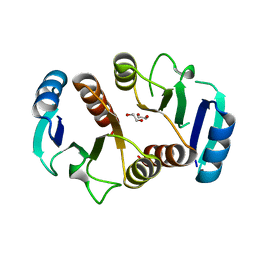 | | 1.6A resolution structure of the ChxR receiver domain from Chlamydia trachomatis | | Descriptor: | 1,2-ETHANEDIOL, Transcriptional regulatory protein | | Authors: | Hickey, J, Lovell, S, Battaile, K.P, Hu, L, Middaugh, C.R, Hefty, P.S. | | Deposit date: | 2011-01-05 | | Release date: | 2011-07-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The atypical response regulator protein ChxR has structural characteristics and dimer interface interactions that are unique within the OmpR/PhoB subfamily.
J.Biol.Chem., 286, 2011
|
|
2NCS
 
 | | NMR assignment and structure of a peptide derived from the membrane proximal external region of HIV-1 gp41 in the presence of dodecylphosphocholine micelles | | Descriptor: | Envelope glycoprotein gp41 | | Authors: | Jimenez, M, Nieva, J.L, Rujas, E, Partida-Hanon, A, Bruix, M. | | Deposit date: | 2016-04-14 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for broad neutralization of HIV-1 through the molecular recognition of 10E8 helical epitope at the membrane interface.
Sci Rep, 6, 2016
|
|
3Q7S
 
 | | 2.1A resolution structure of the ChxR receiver domain containing I3C from Chlamydia trachomatis | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, Transcriptional regulatory protein | | Authors: | Hickey, J, Lovell, S, Battaile, K.P, Hu, L, Middaugh, C.R, Hefty, P.S. | | Deposit date: | 2011-01-05 | | Release date: | 2011-07-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The atypical response regulator protein ChxR has structural characteristics and dimer interface interactions that are unique within the OmpR/PhoB subfamily.
J.Biol.Chem., 286, 2011
|
|
