5M7L
 
 | | Blastochloris viridis photosynthetic reaction center synchrotron structure | | Descriptor: | (2E,6E,10E,14E,18E,22E,26E)-3,7,11,15,19,23,27,31-OCTAMETHYLDOTRIACONTA-2,6,10,14,18,22,26,30-OCTAENYL TRIHYDROGEN DIPHOSPHATE, 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL A, ... | | Authors: | Sharma, A.S, Johansson, L, Dunevall, E, Wahlgren, W.Y, Neutze, R, Katona, G. | | Deposit date: | 2016-10-28 | | Release date: | 2017-02-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Asymmetry in serial femtosecond crystallography data.
Acta Crystallogr A Found Adv, 73, 2017
|
|
6QU9
 
 | | Fab fragment of an antibody that inhibits polymerisation of alpha-1-antitrypsin | | Descriptor: | FAB 4B12 heavy chain, FAB 4B12 light chain, GLYCEROL, ... | | Authors: | Jagger, A.M, Heyer-Chauhan, N, Lomas, D.A, Irving, J.A. | | Deposit date: | 2019-02-26 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis for Z alpha 1 -antitrypsin polymerization in the liver.
Sci Adv, 6, 2020
|
|
5MAI
 
 | | Crystal structure of MELK in complex with an inhibitor | | Descriptor: | 3-[(3~{Z})-3-[[[4-[(dimethylamino)methyl]phenyl]amino]-phenyl-methylidene]-2-oxidanylidene-1~{H}-indol-6-yl]-~{N}-ethyl-prop-2-ynamide, DIMETHYL SULFOXIDE, Maternal embryonic leucine zipper kinase | | Authors: | Canevari, G, Re Depaolini, S, Casale, E, Felder, E, Kuster, B, Heinzlmeir, S. | | Deposit date: | 2016-11-03 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The target landscape of clinical kinase drugs.
Science, 358, 2017
|
|
6UO1
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with mRNA (containing pseudouridine at the first position of the codon) and deacylated A-, P-, and E-site tRNAs at 2.95A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Batool, Z, Dobosz-Bartoszek, M, Polikanov, Y.S. | | Deposit date: | 2019-10-14 | | Release date: | 2019-11-27 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Pseudouridinylation of mRNA coding sequences alters translation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4NH2
 
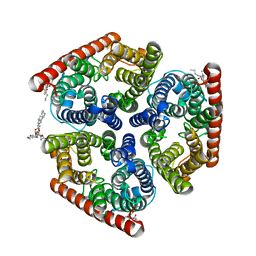 | | Crystal structure of AmtB from E. coli bound to phosphatidylglycerol | | Descriptor: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, Ammonia channel | | Authors: | Laganowsky, A, Reading, E, Allison, T.M, Robinson, C.V. | | Deposit date: | 2013-11-04 | | Release date: | 2014-06-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Membrane proteins bind lipids selectively to modulate their structure and function.
Nature, 510, 2014
|
|
3BGR
 
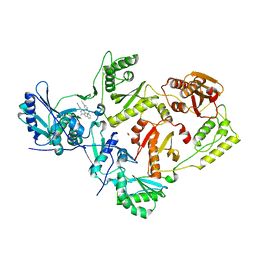 | | Crystal structure of K103N/Y181C mutant HIV-1 reverse transcriptase (RT) in complex with TMC278 (Rilpivirine), a non-nucleoside RT inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, Reverse transcriptase/ribonuclease H, ... | | Authors: | Das, K, Bauman, J.D, Clark Jr, A.D, Shatkin, A.J, Arnold, E. | | Deposit date: | 2007-11-27 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-resolution structures of HIV-1 reverse transcriptase/TMC278 complexes: Strategic flexibility explains potency against resistance mutations.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3TH8
 
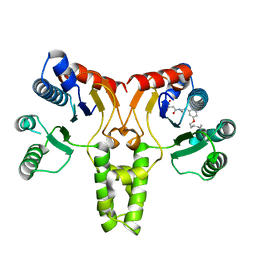 | | Structure of E. coli undecaprenyl diphosphate synthase complexed with BPH-1063 | | Descriptor: | (2Z)-4-({3-[3-(hexyloxy)phenyl]propyl}amino)-2-hydroxy-4-oxobut-2-enoic acid, Undecaprenyl pyrophosphate synthase | | Authors: | Cao, R, Zhu, W, Zhang, Y, Oldfield, E. | | Deposit date: | 2011-08-18 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.114 Å) | | Cite: | HIV-1 Integrase Inhibitor-Inspired Antibacterials Targeting Isoprenoid Biosynthesis.
ACS Med Chem Lett, 3, 2012
|
|
1SFE
 
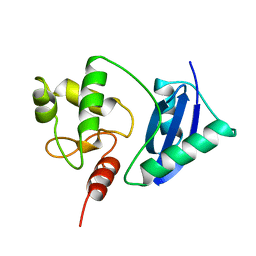 | | ADA O6-METHYLGUANINE-DNA METHYLTRANSFERASE FROM ESCHERICHIA COLI | | Descriptor: | ADA O6-METHYLGUANINE-DNA METHYLTRANSFERASE | | Authors: | Moore, M.H, Gulbis, J.M, Dodson, E.J, Demple, B, Moody, P.C.E. | | Deposit date: | 1996-06-21 | | Release date: | 1996-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a suicidal DNA repair protein: the Ada O6-methylguanine-DNA methyltransferase from E. coli.
EMBO J., 13, 1994
|
|
4DVG
 
 | |
3BRQ
 
 | | Crystal structure of the Escherichia coli transcriptional repressor ascG | | Descriptor: | HTH-type transcriptional regulator ascG, SODIUM ION, SULFATE ION, ... | | Authors: | Singer, A.U, Kagan, O, Evdokimova, E, Osipiuk, J, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-21 | | Release date: | 2008-01-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the E. coli transcriptional repressor ascG.
To be Published
|
|
2ZD1
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (RT) in Complex with TMC278 (Rilpivirine), A Non-nucleoside RT Inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, Reverse transcriptase/ribonuclease H, ... | | Authors: | Das, K, Bauman, J.D, Clark Jr, A.D, Shatkin, A.J, Arnold, E. | | Deposit date: | 2007-11-16 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structures of HIV-1 reverse transcriptase/TMC278 complexes: Strategic flexibility explains potency against resistance mutations.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3ZG4
 
 | | NMR structure of the catalytic domain from E. faecium L,D- transpeptidase | | Descriptor: | ERFK/YBIS/YCFS/YNHG | | Authors: | Lecoq, L, Dubee, V, Triboulet, S, Bougault, C, Hugonnet, J.E, Arthur, M, Simorre, J.P. | | Deposit date: | 2012-12-14 | | Release date: | 2013-04-24 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The Structure of Enterococcus Faecium L,D---Transpeptidase Acylated by Ertapenem Provides Insight Into the Inactivation Mechanism.
Acs Chem.Biol., 8, 2013
|
|
2GBH
 
 | | NMR structure of stem region of helix-35 of 23S E.coli ribosomal RNA (residues 736-760) | | Descriptor: | 5'-R(*(GMP)P*GP*GP*CP*UP*AP*AP*UP*GP*(PSU)P*UP*GP*AP*AP*AP*AP*AP*UP*UP*AP*GP*CP*CP*C)-3' | | Authors: | Bax, A, Boisbouvier, J, Bryce, D, Grishaev, A, Jaroniec, C, Miclet, E, Nikonovicz, E, O'Neil-Cabello, E, Ying, J. | | Deposit date: | 2006-03-10 | | Release date: | 2006-04-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Measurement of five dipolar couplings from a single 3D NMR multiplet applied to the study of RNA dynamics.
J.Am.Chem.Soc., 126, 2004
|
|
5MG0
 
 | | Structure of PAS-GAF fragment of Deinococcus phytochrome by serial femtosecond crystallography | | Descriptor: | 1,2-ETHANEDIOL, 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome, ... | | Authors: | Burgie, E.S, Fuller, F.D, Gul, S, Miller, M.D, Young, I.D, Brewster, A.S, Clinger, J, Aller, P, Braeuer, P, Hutchison, C, Alonso-Mori, R, Kern, J, Yachandra, V.K, Yano, J, Sauter, N.K, Phillips Jr, G.N, Vierstra, R.D, Orville, A.M. | | Deposit date: | 2016-11-20 | | Release date: | 2017-02-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Drop-on-demand sample delivery for studying biocatalysts in action at X-ray free-electron lasers.
Nat. Methods, 14, 2017
|
|
1PS6
 
 | | Crystal structure of E.coli PdxA | | Descriptor: | 4-HYDROXY-L-THREONINE-5-MONOPHOSPHATE, 4-hydroxythreonine-4-phosphate dehydrogenase, ZINC ION | | Authors: | Sivaraman, J, Li, Y, Banks, J, Cane, D.E, Matte, A, Cygler, M. | | Deposit date: | 2003-06-20 | | Release date: | 2003-11-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Escherichia coli PdxA, an Enzyme Involved in the Pyridoxal Phosphate Biosynthesis Pathway
J.Biol.Chem., 278, 2003
|
|
5M7J
 
 | | Blastochloris viridis photosynthetic reaction center structure using best crystal approach | | Descriptor: | (2E,6E,10E,14E,18E,22E,26E)-3,7,11,15,19,23,27,31-OCTAMETHYLDOTRIACONTA-2,6,10,14,18,22,26,30-OCTAENYL TRIHYDROGEN DIPHOSPHATE, 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL A, ... | | Authors: | Sharma, A.S, Johansson, L, Dunevall, E, Wahlgren, W.Y, Neutze, R, Katona, G. | | Deposit date: | 2016-10-28 | | Release date: | 2017-02-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Asymmetry in serial femtosecond crystallography data.
Acta Crystallogr A Found Adv, 73, 2017
|
|
3ZJV
 
 | | Ternary complex of E .coli leucyl-tRNA synthetase, tRNA(Leu) and the benzoxaborole AN3213 in the editing conformation | | Descriptor: | LEUCINE--TRNA LIGASE, TRNALEU5 UAA ISOACCEPTOR | | Authors: | Cusack, S, Palencia, A, Crepin, T, Hernandez, V, Akama, T, Baker, S.J, Bu, W, Feng, L, Freund, Y.R, Liu, L, Meewan, M, Mohan, M, Mao, W, Rock, F.L, Sexton, H, Sheoran, A, Zhang, Y, Zhang, Y, Zhou, Y, Nieman, J.A, Anugula, M.R, Keramane, E.M, Savariraj, K, Reddy, D.S, Sharma, R, Subedi, R, Singh, R, OLeary, A, Simon, N.L, DeMarsh, P.L, Mushtaq, S, Warner, M, Livermore, D.M, Alley, M.R.K, Plattner, J.J. | | Deposit date: | 2013-01-18 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Discovery of a Novel Class of Boron-Based Antibacterials with Activity Against Gram-Negative Bacteria.
Antimicrob.Agents Chemother., 57, 2013
|
|
1PS7
 
 | | Crystal structure of E.coli PdxA | | Descriptor: | 4-hydroxythreonine-4-phosphate dehydrogenase, ZINC ION | | Authors: | Sivaraman, J, Li, Y, Banks, J, Cane, D.E, Matte, A, Cygler, M. | | Deposit date: | 2003-06-20 | | Release date: | 2003-11-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal Structure of Escherichia coli PdxA, an Enzyme Involved in the Pyridoxal Phosphate Biosynthesis Pathway
J.Biol.Chem., 278, 2003
|
|
3ULP
 
 | | Plasmodium falciparum SSB complex with ssDNA | | Descriptor: | DNA (35-MER), Single-strand binding protein | | Authors: | Antony, E, Lohman, T.M, Korolev, S. | | Deposit date: | 2011-11-11 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Plasmodium falciparum SSB Tetramer Wraps Single-Stranded DNA with Similar Topology but Opposite Polarity to E. coli SSB.
J.Mol.Biol., 420, 2012
|
|
6O66
 
 | |
4K2M
 
 | | Crystal structure of ntda from bacillus subtilis in complex with the plp external aldimine adduct with kanosamine-6-phosphate | | Descriptor: | 1,2-ETHANEDIOL, 3-deoxy-3-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-6-O-phosphono-alpha-D-gluco pyranose, ACETATE ION, ... | | Authors: | Van Straaten, K.E, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2013-04-09 | | Release date: | 2013-10-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | The Structure of NtdA, a Sugar Aminotransferase Involved in the Kanosamine Biosynthetic Pathway in Bacillus subtilis, Reveals a New Subclass of Aminotransferases.
J.Biol.Chem., 288, 2013
|
|
1SU1
 
 | | Structural and biochemical characterization of Yfce, a phosphoesterase from E. coli | | Descriptor: | Hypothetical protein yfcE, SULFATE ION, ZINC ION | | Authors: | Miller, D.J, Shuvalova, L, Evdokimova, E, Savchenko, A, Yakunin, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-03-25 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and biochemical characterization of a novel Mn2+-dependent phosphodiesterase encoded by the yfcE gene.
Protein Sci., 16, 2007
|
|
9ARE
 
 | | Rat GluN1-GluN2B NMDA receptor channel in complex with glycine, glutamate, and EU-1622-A, in open-channel conformation | | Descriptor: | GLUTAMIC ACID, GLYCINE, Glutamate receptor ionotropic, ... | | Authors: | Chou, T.-H, Furukawa, H. | | Deposit date: | 2024-02-23 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Molecular mechanism of ligand gating and opening of NMDA receptor.
Nature, 632, 2024
|
|
9ARI
 
 | |
7KQ9
 
 | |
