1NRK
 
 | |
1NRL
 
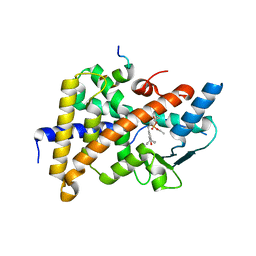 | | Crystal Structure of the human PXR-LBD in complex with an SRC-1 coactivator peptide and SR12813 | | Descriptor: | Nuclear Receptor Coactivator 1 isoform 3, Orphan nuclear receptor PXR, [2-(3,5-DI-TERT-BUTYL-4-HYDROXY-PHENYL)-1-(DIETHOXY-PHOSPHORYL)-VINYL]-PHOSPHONIC ACID DIETHLYL ESTER | | Authors: | Watkins, R.E, Davis-Searles, P.R, Lambert, M.H, Redinbo, M.R. | | Deposit date: | 2003-01-25 | | Release date: | 2003-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Coactivator binding promotes the specific interaction between ligand and the pregnane X receptor
J.Mol.Biol., 331, 2003
|
|
1NRM
 
 | |
1NRN
 
 | |
1NRO
 
 | |
1NRP
 
 | |
1NRQ
 
 | |
1NRR
 
 | |
1NRS
 
 | |
1NRU
 
 | |
1NRV
 
 | | Crystal structure of the SH2 domain of Grb10 | | Descriptor: | Growth factor receptor-bound protein 10 | | Authors: | Stein, E.G, Hubbard, S.R. | | Deposit date: | 2003-01-25 | | Release date: | 2003-04-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for dimerization of the Grb10 Src homology 2 domain. Implications for ligand specificity.
J.Biol.Chem., 278, 2003
|
|
1NRW
 
 | | The structure of a HALOACID DEHALOGENASE-LIKE HYDROLASE FROM B. SUBTILIS | | Descriptor: | CALCIUM ION, PHOSPHATE ION, hypothetical protein, ... | | Authors: | Cuff, M.E, Kim, Y, Zhang, R, Joachimiak, A, Collart, F, Quartey, P, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-25 | | Release date: | 2003-07-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of a HALOACID DEHALOGENASE-LIKE HYDROLASE FROM B. SUBTILIS
To be Published
|
|
1NRX
 
 | | Crystal structure of 3-dehydroquinate synthase (DHQS) in complex with ZN2+ and NAD | | Descriptor: | 3-dehydroquinate synthase, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Nichols, C.E, Ren, J, Lamb, H.K, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2003-01-26 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Ligand-induced Conformational Changes and a Mechanism for Domain Closure in Aspergillus nidulans Dehydroquinate Synthase
J.MOL.BIOL., 327, 2003
|
|
1NRZ
 
 | |
1NS0
 
 | |
1NS1
 
 | |
1NS2
 
 | |
1NS3
 
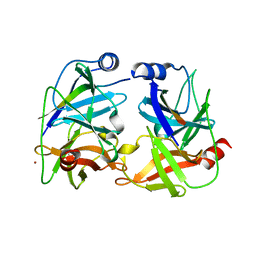 | | STRUCTURE OF HCV PROTEASE (BK STRAIN) | | Descriptor: | NS3 PROTEASE, NS4A PEPTIDE, ZINC ION | | Authors: | Yan, Y, Munshi, S, Chen, Z. | | Deposit date: | 1997-04-05 | | Release date: | 1998-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Complex of NS3 protease and NS4A peptide of BK strain hepatitis C virus: a 2.2 A resolution structure in a hexagonal crystal form.
Protein Sci., 7, 1998
|
|
1NS4
 
 | |
1NS5
 
 | | X-RAY STRUCTURE OF YBEA FROM E.COLI. NORTHEAST STRUCTURAL GENOMICS RESEARCH CONSORTIUM (NESG) TARGET ER45 | | Descriptor: | Hypothetical protein ybeA | | Authors: | Benach, J, Shen, J, Rost, B, Xiao, R, Acton, T, Montelione, G, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-01-27 | | Release date: | 2003-02-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structure of YBEA from E. coli
To be Published
|
|
1NS6
 
 | |
1NS7
 
 | |
1NS8
 
 | |
1NS9
 
 | |
1NSA
 
 | |
