4J8X
 
 | |
9C0I
 
 | |
2DK5
 
 | | Solution structure of Winged-Helix domain in RNA polymerase III 39KDa polypeptide | | Descriptor: | DNA-directed RNA polymerase III 39 kDa polypeptide | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-06 | | Release date: | 2006-10-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Winged-Helix domain in RNA polymerase III 39KDa polypeptide
To be Published
|
|
5KDD
 
 | | Apo-structure of humanised RadA-mutant humRadA22 | | Descriptor: | DNA repair and recombination protein RadA, SULFATE ION | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-06-08 | | Release date: | 2016-10-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
3V30
 
 | | Crystal Structure of the Peptide Bound Complex of the Ankyrin Repeat Domains of Human RFXANK | | Descriptor: | DNA-binding protein RFX5, DNA-binding protein RFXANK | | Authors: | Lam, R, Xu, C, Bian, C.B, Kania, J, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-12-12 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Sequence-Specific Recognition of a PxLPxI/L Motif by an Ankyrin Repeat Tumbler Lock.
Sci.Signal., 5, 2012
|
|
5B33
 
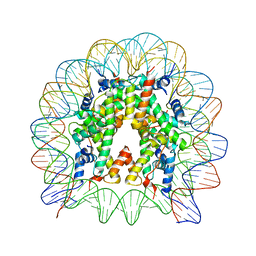 | | The crystal structure of the H2AZ nucleosome with H3.3. | | Descriptor: | DNA (146-MER), Histone H2A.Z, Histone H2B type 1-J, ... | | Authors: | Horikoshi, N, Taguchi, H, Arimura, Y, Kurumizaka, H. | | Deposit date: | 2016-02-08 | | Release date: | 2016-08-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.925 Å) | | Cite: | Crystal structures of heterotypic nucleosomes containing histones H2A.Z and H2A.
Open Biology, 6, 2016
|
|
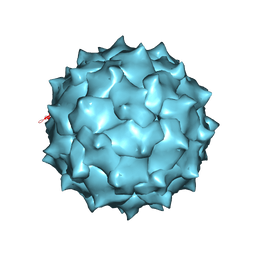
4MGU
 
 | |
6ED9
 
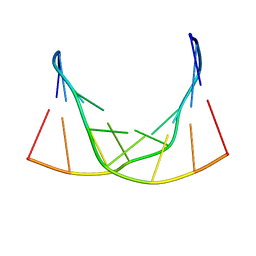 | |
2L0I
 
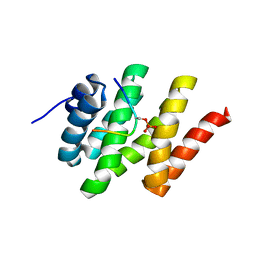 | | Solution structure of Rtt103 CTD-interacting domain bound to a Ser2 phosphorylated CTD peptide | | Descriptor: | DNA-directed RNA polymerase, Regulator of Ty1 transposition protein 103 | | Authors: | Lunde, B.M, Reichow, S.L, Kim, M, Suh, H, Leeper, T.C, Yang, F, Mutschler, H, Buratowski, S, Meinhart, A, Varani, G. | | Deposit date: | 2010-07-06 | | Release date: | 2010-09-08 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Cooperative interaction of transcription termination factors with the RNA polymerase II C-terminal domain.
Nat.Struct.Mol.Biol., 17, 2010
|
|
7Y3P
 
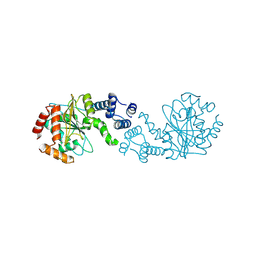 | |
5XP8
 
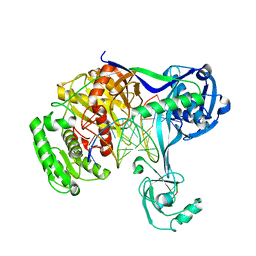 | | Crystal structure of T. thermophilus Argonaute protein complexed with a bulge 4A5 on the guide strand | | Descriptor: | DNA (5'-D(*AP*GP*T)-3'), DNA (5'-D(*CP*AP*AP*CP*C*AP*TP*AP*CP*TP*AP*CP*CP*TP*CP*G)-3'), DNA (5'-D(P*TP*GP*AP*GP*AP*GP*TP*AP*GP*TP*AP*GP*GP*TP*TP*GP*T)-3'), ... | | Authors: | Sheng, G, Gogakos, T, Wang, J, Zhao, H, Serganov, A, Juranek, S, Tuschl, T, Patel, J.D, Wang, Y. | | Deposit date: | 2017-06-01 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure/cleavage-based insights into helical perturbations at bulge sites within T. thermophilus Argonaute silencing complexes.
Nucleic Acids Res., 45, 2017
|
|
2LNB
 
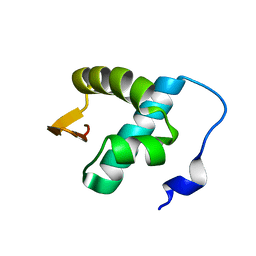 | | Solution NMR structure of N-terminal domain (6-74) of human ZBP1 protein, Northeast Structural Genomics Consortium Target HR8174A. | | Descriptor: | Z-DNA-binding protein 1 | | Authors: | Yang, Y, Ramelot, T.A, Hamilton, K, Kohan, E, Wang, D, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of N-terminal domain (6-74) of human ZBP1 protein, Northeast Structural Genomics Consortium Target HR8174A
To be Published
|
|
5XQ2
 
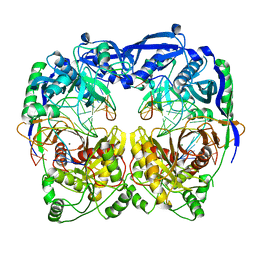 | | Crystal structure of T. thermophilus Argonaute protein complexed with a bulge 5A6 on the guide strand | | Descriptor: | DNA (5'-D(*AP*CP*AP*AP*CP*CP*TP*AP*CP*TP*AP*CP*CP*TP*CP*G)-3'), DNA (5'-D(*AP*GP*T)-3'), DNA (5'-D(P*TP*GP*AP*AP*GP*AP*TP*AP*GP*TP*AP*GP*GP*TP*TP*GP*T)-3'), ... | | Authors: | Sheng, G, Gogakos, T, Wang, J, Zhao, H, Serganov, A, Juranek, S, Tuschl, T, Patel, D, Wang, Y. | | Deposit date: | 2017-06-06 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Structure/cleavage-based insights into helical perturbations at bulge sites within T. thermophilus Argonaute silencing complexes
Nucleic Acids Res., 45, 2017
|
|
1UD9
 
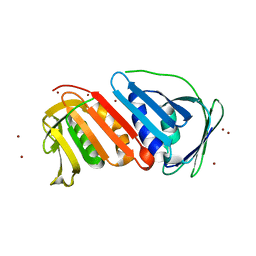 | | Crystal Structure of Proliferating Cell Nuclear Antigen (PCNA) Homolog From Sulfolobus tokodaii | | Descriptor: | DNA polymerase sliding clamp A, ZINC ION | | Authors: | Tanabe, E, Yasutake, Y, Tanaka, Y, Yao, M, Tsumoto, K, Kumagai, I, Tanaka, I. | | Deposit date: | 2003-04-28 | | Release date: | 2004-06-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Structure of Proliferating Cell Nuclear Antigen (PCNA) Homolog From Sulfolobus tokodaii
To be published
|
|
6KMS
 
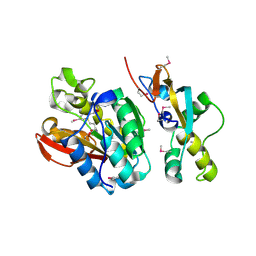 | | Crystal structure of human N6amt1-Trm112 in complex with SAM (space group I422) | | Descriptor: | Methyltransferase N6AMT1, Multifunctional methyltransferase subunit TRM112-like protein, S-ADENOSYLMETHIONINE | | Authors: | Li, W.J, Shi, Y, Zhang, T.L, Ye, J, Ding, J.P. | | Deposit date: | 2019-08-01 | | Release date: | 2019-09-18 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insight into human N6amt1-Trm112 complex functioning as a protein methyltransferase.
Cell Discov, 5, 2019
|
|
6KMR
 
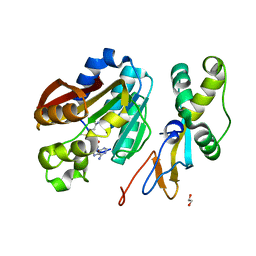 | | Crystal structure of human N6amt1-Trm112 in complex with SAM (space group P6122) | | Descriptor: | 1,2-ETHANEDIOL, Methyltransferase N6AMT1, Multifunctional methyltransferase subunit TRM112-like protein, ... | | Authors: | Li, W.J, Shi, Y, Zhang, T.L, Ye, J, Ding, J.P. | | Deposit date: | 2019-08-01 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight into human N6amt1-Trm112 complex functioning as a protein methyltransferase.
Cell Discov, 5, 2019
|
|
1DOU
 
 | | MONOVALENT CATIONS SEQUESTER WITHIN THE A-TRACT MINOR GROOVE OF [D(CGCGAATTCGCG)]2 | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Woods, K.K, McFail-Isom, L, Sines, C.C, Howerton, S.B, Stephens, R.K, Williams, L.D. | | Deposit date: | 1999-12-21 | | Release date: | 1999-12-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Monovalent Cations Sequester within the A-Tract Minor Groove of [d(CGCGAATTCGCG)]2
J.Am.Chem.Soc., 122, 2000
|
|
2L42
 
 | |
5ZWU
 
 | |
8PMB
 
 | |
6SW3
 
 | |
8R6D
 
 | |
5XOU
 
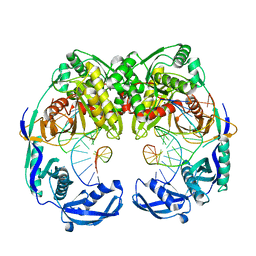 | | Crystal structure of T. thermophilus Argonaute protein complexed with a bulge 7T8 on the guide strand | | Descriptor: | DNA (5'-D(*AP*CP*AP*AP*CP*CP*TP*AP*CP*TP*AP*CP*CP*TP*CP*G)-3'), DNA (5'-D(P*TP*GP*AP*GP*GP*TP*AP*TP*GP*GP*TP*TP*GP*T)-3'), MAGNESIUM ION, ... | | Authors: | Sheng, G, Wang, J, Zhao, H, Wang, Y. | | Deposit date: | 2017-05-31 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structure/cleavage-based insights into helical perturbations at bulge sites within T. thermophilus Argonaute silencing complexes
Nucleic Acids Res., 45, 2017
|
|
6O1Z
 
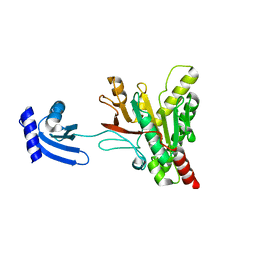 | | Structure of pCW3 conjugation coupling protein TcpA hexagonal crystal form | | Descriptor: | DNA translocase coupling protein | | Authors: | Traore, D.A.K, Ahktar, N, Torres, V.T, Adams, V, Coulibaly, F, Panjikar, S, Caradoc-Davies, T.T, Rood, J.I, Whisstock, J.C. | | Deposit date: | 2019-02-22 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of pCW3 conjugation coupling protein TcpA
To be published
|
|
6NN6
 
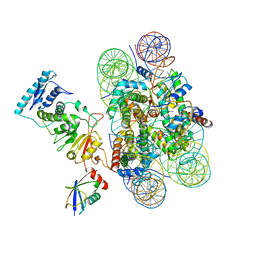 | | Structure of Dot1L-H2BK120ub nucleosome complex | | Descriptor: | DNA (145-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Anderson, C.J, Baird, M.R, Hsu, A, Barbour, E.H, Koyama, Y, Borgnia, M.J, McGinty, R.K. | | Deposit date: | 2019-01-14 | | Release date: | 2019-02-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Basis for Recognition of Ubiquitylated Nucleosome by Dot1L Methyltransferase.
Cell Rep, 26, 2019
|
|
