3SUA
 
 | | Crystal structure of the intracellular domain of Plexin-B1 in complex with Rac1 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Plexin-B1, ... | | Authors: | Bell, C.H, Aricescu, A.R, Jones, E.Y, Siebold, C. | | Deposit date: | 2011-07-11 | | Release date: | 2011-09-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.39 Å) | | Cite: | A Dual Binding Mode for RhoGTPases in Plexin Signalling.
Plos Biol., 9, 2011
|
|
3SU8
 
 | | Crystal structure of a truncated intracellular domain of Plexin-B1 in complex with Rac1 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Plexin-B1, ... | | Authors: | Bell, C.H, Aricescu, A.R, Jones, E.Y, Siebold, C. | | Deposit date: | 2011-07-11 | | Release date: | 2011-09-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A Dual Binding Mode for RhoGTPases in Plexin Signalling.
Plos Biol., 9, 2011
|
|
6TGC
 
 | | CryoEM structure of the ternary DOCK2-ELMO1-RAC1 complex. | | Descriptor: | Dedicator of cytokinesis protein 2, Engulfment and cell motility protein 1, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Chang, L, Yang, J, Chang, J.H, Zhang, Z, Boland, A, McLaughlin, S.H, Abu-Thuraia, A, Killoran, R.C, Smith, M.J, Cote, J.F, Barford, D. | | Deposit date: | 2019-11-15 | | Release date: | 2020-07-29 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of the DOCK2-ELMO1 complex provides insights into regulation of the auto-inhibited state.
Nat Commun, 11, 2020
|
|
8JVJ
 
 | | Structure of human TRPV4 with antagonist A2 and RhoA | | Descriptor: | Transforming protein RhoA, Transient receptor potential cation channel subfamily V member 4,3C-GFP, [6-[[4-(2,4-dimethyl-1,3-thiazol-5-yl)-1,3-thiazol-2-yl]amino]pyridin-3-yl]-[(1~{S},5~{R})-3-[5-(trifluoromethyl)pyrimidin-2-yl]-3,8-diazabicyclo[3.2.1]octan-8-yl]methanone | | Authors: | Fan, J, Lei, X. | | Deposit date: | 2023-06-28 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structural Pharmacology of TRPV4 Antagonists.
Adv Sci, 11, 2024
|
|
6HUW
 
 | |
2WM9
 
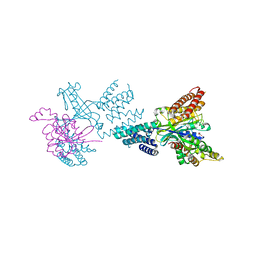 | | Structure of the complex between DOCK9 and Cdc42. | | Descriptor: | CELL DIVISION CONTROL PROTEIN 42 HOMOLOG, DEDICATOR OF CYTOKINESIS PROTEIN 9, GLYCEROL | | Authors: | Yang, J, Roe, S.M, Barford, D. | | Deposit date: | 2009-06-30 | | Release date: | 2009-09-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Activation of Rho Gtpases by Dock Exchange Factors is Mediated by a Nucleotide Sensor.
Science, 325, 2009
|
|
3MSX
 
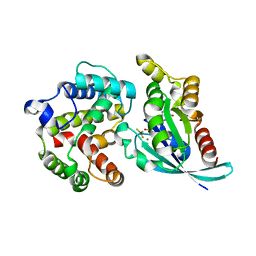 | | Crystal structure of RhoA.GDP.MgF3 in complex with GAP domain of ArhGAP20 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Rho GTPase-activating protein 20, ... | | Authors: | Utepbergenov, D, Cooper, D.R, Derewenda, U, Derewenda, Z.S. | | Deposit date: | 2010-04-29 | | Release date: | 2011-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mechanism of molecular specificity of RhoGAP domains towards small GTPases of RhoA family.
To be Published
|
|
2WMO
 
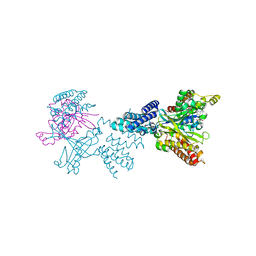 | | Structure of the complex between DOCK9 and Cdc42. | | Descriptor: | CELL DIVISION CONTROL PROTEIN 42 HOMOLOG, DEDICATOR OF CYTOKINESIS PROTEIN 9, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yang, J, Roe, S.M, Barford, D. | | Deposit date: | 2009-07-02 | | Release date: | 2009-09-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Activation of Rho Gtpases by Dock Exchange Factors is Mediated by a Nucleotide Sensor.
Science, 325, 2009
|
|
3T06
 
 | |
2X13
 
 | | The catalytically active fully closed conformation of human phosphoglycerate kinase in complex with ADP and 3phosphoglycerate | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bowler, M.W, Cliff, M.J, Marston, J.P.M, Baxter, N.J, Hownslow, A.M.H, Varga, A.V, Szabo, J, Vas, M, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-12-21 | | Release date: | 2010-12-29 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Metal fluorides-multi-functional tools for the study of phosphoryl transfer enzymes, a practical guide.
Structure, 2024
|
|
2X15
 
 | | The catalytically active fully closed conformation of human phosphoglycerate kinase in complex with ADP and 1,3- bisphosphoglycerate | | Descriptor: | 1,3-BISPHOSPHOGLYCERIC ACID, 3-PHOSPHOGLYCERIC ACID, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Bowler, M.W, Cliff, M.J, Marston, J.P.M, Baxter, N.J, Hounslow, A.M.H, Varga, A.V, Szabo, J, Vas, M, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-12-21 | | Release date: | 2011-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of Human Phosphoglycerate Kinase in its Fully Active Conformation in Complex with Ground State Analoges
To be Published
|
|
3SBE
 
 | | Crystal structure of RAC1 P29S mutant | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Ha, B.H, Boggon, T.J. | | Deposit date: | 2011-06-03 | | Release date: | 2012-07-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma.
Nat.Genet., 44, 2012
|
|
2QVX
 
 | | 4-Chlorobenzoyl-CoA Ligase/Synthetase, I303G mutation, bound to 3-Chlorobenzoate | | Descriptor: | 3-chlorobenzoate, 4-Chlorobenzoate CoA Ligase | | Authors: | Wu, R, Reger, A.S, Cao, J, Gulick, A.M, Dunaway-Mariano, D. | | Deposit date: | 2007-08-09 | | Release date: | 2007-12-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Rational redesign of the 4-chlorobenzoate binding site of 4-chlorobenzoate: coenzyme a ligase for expanded substrate range.
Biochemistry, 46, 2007
|
|
3TH5
 
 | | Crystal structure of wild-type RAC1 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Ha, B.H, Boggon, T.J. | | Deposit date: | 2011-08-18 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma.
Nat.Genet., 44, 2012
|
|
3U17
 
 | | Structure of BasE N-terminal domain from Acinetobacter baumannii bound to 6-(p-benzoyl)phenyl-1-(pyridin-4-ylmethyl)-1H-pyrazolo[3,4-b]pyridine-4-carboxylic acid | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 6-(4-benzoylphenyl)-1-(pyridin-4-ylmethyl)-1H-pyrazolo[3,4-b]pyridine-4-carboxylic acid, ... | | Authors: | Gulick, A.M, Drake, E.J, Aldrich, C.C, Neres, J. | | Deposit date: | 2011-09-29 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Non-nucleoside inhibitors of BasE, an adenylating enzyme in the siderophore biosynthetic pathway of the opportunistic pathogen Acinetobacter baumannii.
J.Med.Chem., 56, 2013
|
|
7SF8
 
 | | GPR56 (ADGRG1) 7TM domain bound to tethered agonist in complex with G protein heterotrimer | | Descriptor: | G protein subunit 13 (Gi2-mini-G13 chimera), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Barros-Alvarez, X, Panova, O, Skiniotis, G. | | Deposit date: | 2021-10-03 | | Release date: | 2022-04-27 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The tethered peptide activation mechanism of adhesion GPCRs.
Nature, 604, 2022
|
|
7SJ4
 
 | | Human Trio residues 1284-1959 in complex with Rac1 | | Descriptor: | Ras-related C3 botulinum toxin substrate 1, Triple functional domain protein | | Authors: | Chen, C.-L, Ravala, S.K, Bandekar, S.J, Cash, J, Tesmer, J.J.G. | | Deposit date: | 2021-10-15 | | Release date: | 2022-07-06 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural/functional studies of Trio provide insights into its configuration and show that conserved linker elements enhance its activity for Rac1.
J.Biol.Chem., 298, 2022
|
|
7KCP
 
 | |
5ZV6
 
 | |
7L3Q
 
 | |
6YQM
 
 | | Human histidine triad nucleotide-binding protein 1 (hHINT1) complexed with dGMP and refined to 1.02 A | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, Histidine triad nucleotide-binding protein 1 | | Authors: | Dolot, R.D, Seda, A, Nawrot, B.C. | | Deposit date: | 2020-04-17 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Biochemical, crystallographic and biophysical characterization of histidine triad nucleotide-binding protein 2 with different ligands including a non-hydrolyzable analog of Ap4A.
Biochim Biophys Acta Gen Subj, 1865, 2021
|
|
1NV4
 
 | | Fructose-1,6-Bisphosphatase Complex with Magnesium, Fructose-6-Phosphate, Phosphate, EDTA and Thallium (1 mM) | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase, MAGNESIUM ION, ... | | Authors: | Choe, J, Iancu, C.V, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2003-02-02 | | Release date: | 2003-07-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Interaction of Tl+ with product complexes of fructose-1,6-bisphosphatase
J.BIOL.CHEM., 278, 2003
|
|
3FI3
 
 | | Crystal structure of JNK3 with indazole inhibitor, SR-3737 | | Descriptor: | 1,2-ETHANEDIOL, 3-{5-[(2-fluorophenyl)amino]-1H-indazol-1-yl}-N-(3,4,5-trimethoxyphenyl)benzamide, Mitogen-activated protein kinase 10 | | Authors: | Habel, J.E, Duckett, D, LoGrasso, P. | | Deposit date: | 2008-12-10 | | Release date: | 2009-03-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-activity relationships and X-ray structures describing the selectivity of aminopyrazole inhibitors for c-Jun N-terminal kinase 3 (JNK3) over p38.
J.Biol.Chem., 284, 2009
|
|
3FI2
 
 | | Crystal structure of JNK3 with amino-pyrazole inhibitor, SR-3451 | | Descriptor: | 1,2-ETHANEDIOL, 3-{4-[(phenylcarbamoyl)amino]-1H-pyrazol-1-yl}-N-(3,4,5-trimethoxyphenyl)benzamide, Mitogen-activated protein kinase 10 | | Authors: | Habel, J.E. | | Deposit date: | 2008-12-10 | | Release date: | 2009-03-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure-activity relationships and X-ray structures describing the selectivity of aminopyrazole inhibitors for c-Jun N-terminal kinase 3 (JNK3) over p38.
J.Biol.Chem., 284, 2009
|
|
5YVB
 
 | | Structure of CaMKK2 in complex with CKI-011 | | Descriptor: | (3Z)-5-chloro-3-[(1-methyl-1H-pyrazol-4-yl)methylidene]-1,3-dihydro-2H-indol-2-one, CHLORIDE ION, Calcium/calmodulin-dependent protein kinase kinase 2, ... | | Authors: | Niwa, H, Handa, N, Yokoyama, S. | | Deposit date: | 2017-11-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Protein ligand interaction analysis against new CaMKK2 inhibitors by use of X-ray crystallography and the fragment molecular orbital (FMO) method.
J.Mol.Graph.Model., 99, 2020
|
|
