4C39
 
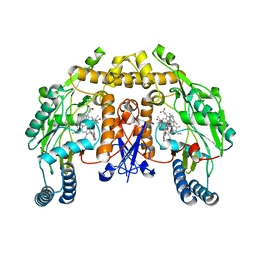 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 6-((((3S, 5R)-5-(((6-amino-4-methylpyridin-2-yl)methoxy) methyl)pyrrolidin-3-yl)oxy)methyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-((((3S, 5R)-5-(((6-amino-4-methylpyridin-2-yl)methoxy)methyl)pyrrolidin-3-yl)oxy)methyl)-4-methylpyridin-2-amine, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2013-08-22 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | An Accessible Chiral Linker to Enhance Potency and Selectivity of Neuronal Nitric Oxide Synthase Inhibitors.
Acs Med.Chem.Lett., 5, 2014
|
|
6LVW
 
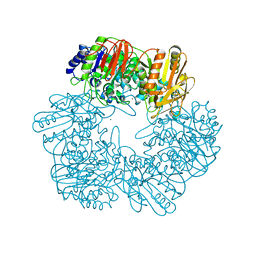 | |
2LFG
 
 | | Solution structure of the human prolactin receptor ecd domain d2 | | Descriptor: | Prolactin receptor | | Authors: | Dagil, R, Knudsen, M.J, Kragelund, B.B, O'Shea, C, Teilum, K. | | Deposit date: | 2011-06-30 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The WSXWS Motif in Cytokine Receptors Is a Molecular Switch Involved in Receptor Activation: Insight from Structures of the Prolactin Receptor
Structure, 20, 2012
|
|
6RQJ
 
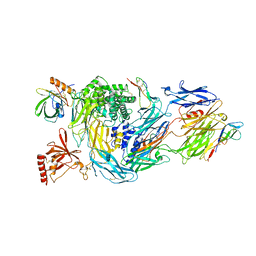 | | Structure of human complement C5 complexed with tick inhibitors OmCI, RaCI1 and CirpT1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C5, Complement inhibitor, ... | | Authors: | Reichhardt, M.P, Johnson, S, Lea, S.M. | | Deposit date: | 2019-05-15 | | Release date: | 2020-01-08 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | An inhibitor of complement C5 provides structural insights into activation.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6LMS
 
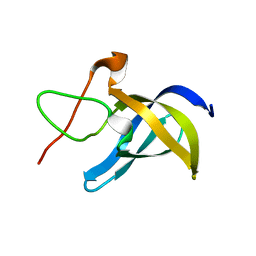 | |
4CAC
 
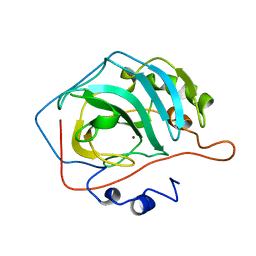 | | REFINED STRUCTURE OF HUMAN CARBONIC ANHYDRASE II AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | CARBONIC ANHYDRASE FORM C, ZINC ION | | Authors: | Lindahl, M, Habash, D, Harrop, S, Helliwell, D.R, Liljas, A. | | Deposit date: | 1991-09-05 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Refined structure of human carbonic anhydrase II at 2.0 A resolution.
Proteins, 4, 1988
|
|
6LR2
 
 | |
6RT4
 
 | |
6LYY
 
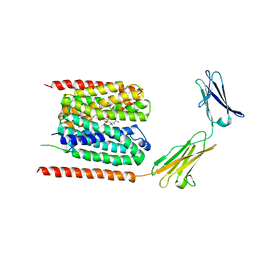 | | Cryo-EM structure of the human MCT1/Basigin-2 complex in the presence of anti-cancer drug candidate AZD3965 in the outward-open conformation. | | Descriptor: | 3-methyl-5-[[(4~{R})-4-methyl-4-oxidanyl-1,2-oxazolidin-2-yl]carbonyl]-6-[[5-methyl-3-(trifluoromethyl)-1~{H}-pyrazol-4-yl]methyl]-1-propan-2-yl-thieno[2,3-d]pyrimidine-2,4-dione, Basigin, Monocarboxylate transporter 1 | | Authors: | Wang, N, Jiang, X, Zhang, S, Zhu, A, Yuan, Y, Lei, J, Yan, C. | | Deposit date: | 2020-02-16 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of human monocarboxylate transporter 1 inhibition by anti-cancer drug candidates.
Cell, 184, 2021
|
|
6RU1
 
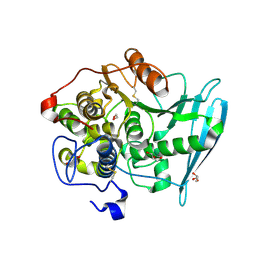 | | Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant in complex with the aldouronic acid Um4X | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, ... | | Authors: | Ernst, H.A, Mosbech, C, Langkilde, A, Westh, P, Meyer, A, Agger, J.W, Larsen, S. | | Deposit date: | 2019-05-27 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | The structural basis of fungal glucuronoyl esterase activity on natural substrates.
Nat Commun, 11, 2020
|
|
3EVE
 
 | |
3E6N
 
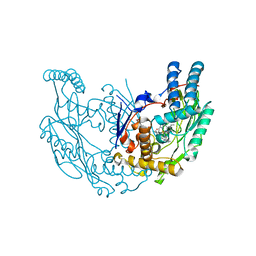 | | Structure of murine INOS oxygenase domain with inhibitor AR-C125813 | | Descriptor: | 4-METHYL-6-PROPYLPYRIDIN-2-AMINE, 5,6,7,8-TETRAHYDROBIOPTERIN, Nitric oxide synthase, ... | | Authors: | Garcin, E.D, Arvai, A.S, Rosenfeld, R.J, Kroeger, M.D, Crane, B.R, Andersson, G, Andrews, G, Hamley, P.J, Mallinder, P.R, Nicholls, D.J, St-Gallay, S.A, Tinker, A.C, Gensmantel, N.P, Mete, A, Cheshire, D.R, Connolly, S, Stueh, D.J, Aberg, A, Wallace, A.V, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2008-08-15 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase.
Nat.Chem.Biol., 4, 2008
|
|
6RQE
 
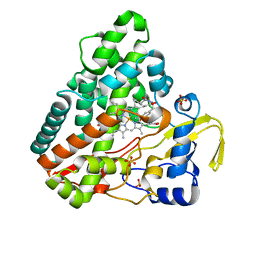 | | CYP121 in complex with 3-acetylene dicyclotyrosine | | Descriptor: | 3-acetylene dicyclotyrosine, Mycocyclosin synthase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Poddar, H, Levy, C. | | Deposit date: | 2019-05-15 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structure-Activity Relationships ofcyclo(l-Tyrosyl-l-tyrosine) Derivatives Binding toMycobacterium tuberculosisCYP121: Iodinated Analogues Promote Shift to High-Spin Adduct.
J.Med.Chem., 62, 2019
|
|
2LAY
 
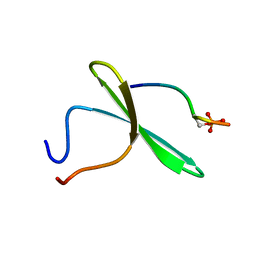 | | Structure of the first WW domain of human YAP in complex with a phosphorylated human Smad1 derived peptide | | Descriptor: | Mothers against decapentaplegic homolog 1, Yorkie homolog | | Authors: | Macias, M.J, Aragon, E, Goerner, N, Zaromytidou, A, Xi, Q, Escobedo, A, Massague, J. | | Deposit date: | 2011-03-22 | | Release date: | 2011-07-06 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | A Smad action turnover switch operated by WW domain readers of a phosphoserine code.
Genes Dev., 25, 2011
|
|
6RQO
 
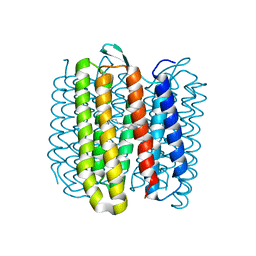 | | Steady-state-SMX activated state structure of bacteriorhodopsin | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Weinert, T, Skopintsev, P, James, D, Kekilli, D, Furrer, A, Bruenle, S, Mous, S, Nogly, P, Standfuss, J. | | Deposit date: | 2019-05-16 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Proton uptake mechanism in bacteriorhodopsin captured by serial synchrotron crystallography.
Science, 365, 2019
|
|
4CBU
 
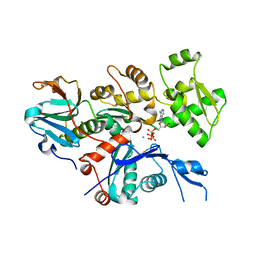 | | Crystal structure of Plasmodium falciparum actin I | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-1, CALCIUM ION, ... | | Authors: | Vahokoski, J, Bhargav, S.P, Desfosses, A, Andreadaki, M, Kumpula, E.P, Ignatev, A, Munico Martinez, S, Lepper, S, Frischknecht, F, Siden-Kiamos, I, Sachse, C, Kursula, I. | | Deposit date: | 2013-10-16 | | Release date: | 2014-04-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Differences Explain Diverse Functions of Plasmodium Actins.
Plos Pathog., 10, 2014
|
|
3E65
 
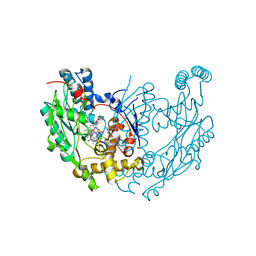 | | Murine INOS dimer with HEME, pterin and inhibitor AR-C120011 | | Descriptor: | 1-[4-(AMINOMETHYL)BENZOYL]-5'-FLUORO-1'H-SPIRO[PIPERIDINE-4,2'-QUINAZOLIN]-4'-AMINE, 5,6,7,8-TETRAHYDROBIOPTERIN, Nitric oxide synthase, ... | | Authors: | Rosenfeld, R.J, Garcin, E.D, Getzoff, E.D. | | Deposit date: | 2008-08-14 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase.
Nat.Chem.Biol., 4, 2008
|
|
2LB1
 
 | | Structure of the second domain of human Smurf1 in complex with a human Smad1 derived peptide | | Descriptor: | E3 ubiquitin-protein ligase SMURF1, Mothers against decapentaplegic homolog 1 | | Authors: | Macias, M.J, Aragon, E, Goerner, N, Zaromytidou, A, Xi, Q, Escobedo, A, Massague, J. | | Deposit date: | 2011-03-22 | | Release date: | 2011-07-06 | | Last modified: | 2016-04-27 | | Method: | SOLUTION NMR | | Cite: | A Smad action turnover switch operated by WW domain readers of a phosphoserine code.
Genes Dev., 25, 2011
|
|
3EAM
 
 | | An open-pore structure of a bacterial pentameric ligand-gated ion channel | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, DODECYL-BETA-D-MALTOSIDE, Glr4197 protein | | Authors: | Bocquet, N, Nury, H, Baaden, M, Le Poupon, C, Changeux, J.P, Delarue, M, Corringer, P.J. | | Deposit date: | 2008-08-26 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation.
Nature, 457, 2009
|
|
3EAI
 
 | | Structure of inhibited murine iNOS oxygenase domain | | Descriptor: | 4-({4-[(4-methoxypyridin-2-yl)amino]piperidin-1-yl}carbonyl)benzonitrile, 5,6,7,8-TETRAHYDROBIOPTERIN, Nitric oxide synthase, ... | | Authors: | Garcin, E.D, Arvai, A.S, Rosenfeld, R.J, Kroeger, M.D, Crane, B.R, Andersson, G, Andrews, G, Hamley, P.J, Mallinder, P.R, Nicholls, D.J, St-Gallay, S.A, Tinker, A.C, Gensmantel, N.P, Mete, A, Cheshire, D.R, Connolly, S, Stuehr, D.J, Aberg, A, Wallace, A.V, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2008-08-25 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase.
Nat.Chem.Biol., 4, 2008
|
|
3EBA
 
 | | CAbHul6 FGLW mutant (humanized) in complex with human lysozyme | | Descriptor: | CAbHul6, Lysozyme C, SULFATE ION | | Authors: | Loris, R, Vincke, C, Saerens, D, Martinez-Rodriguez, S, Muyldermans, S, Conrath, K. | | Deposit date: | 2008-08-27 | | Release date: | 2008-12-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | General Strategy to Humanize a Camelid Single-domain Antibody and Identification of a Universal Humanized Nanobody Scaffold
J.Biol.Chem., 284, 2009
|
|
6LLZ
 
 | |
3EDE
 
 | |
2LP8
 
 | | SOLUTION STRUCTURE OF AN APOPTOSIS ACTIVATING PHOTOSWITCHABLE BAK PEPTIDE BOUND to BCL-XL | | Descriptor: | 3,3'-(E)-diazene-1,2-diylbis{6-[(chloroacetyl)amino]benzenesulfonic acid}, Bcl-2 homologous antagonist/killer, Bcl-2-like protein 1 | | Authors: | Wysoczanski, P, Mart, R.J, Loveridge, J.E, Williams, C, Whittaker, S.B.-M, Crump, M.P, Allemann, R.K. | | Deposit date: | 2012-02-03 | | Release date: | 2012-04-18 | | Last modified: | 2021-08-18 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of a Photoswitchable Apoptosis Activating Bak Peptide Bound to Bcl-x(L).
J.Am.Chem.Soc., 134, 2012
|
|
6RZ8
 
 | | Crystal structure of the human cysteinyl leukotriene receptor 2 in complex with ONO-2080365 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2~{S})-8-[[4-[4-[2,3-bis(fluoranyl)phenoxy]butoxy]-2-fluoranyl-phenyl]carbonylamino]-4-(4-oxidanyl-4-oxidanylidene-but yl)-2,3-dihydro-1,4-benzoxazine-2-carboxylic acid, Cysteinyl leukotriene receptor 2,Soluble cytochrome b562,Cysteinyl leukotriene receptor 2, ... | | Authors: | Gusach, A, Luginina, A, Marin, E, Brouillette, R.L, Besserer-Offroy, E, Longpre, J.M, Ishchenko, A, Popov, P, Fujimoto, T, Maruyama, T, Stauch, B, Ergasheva, M, Romanovskaya, D, Stepko, A, Kovalev, K, Shevtsov, M, Gordeliy, V, Han, G.W, Sarret, P, Katritch, V, Borshchevskiy, V, Mishin, A, Cherezov, V. | | Deposit date: | 2019-06-12 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of ligand selectivity and disease mutations in cysteinyl leukotriene receptors.
Nat Commun, 10, 2019
|
|
