8TV7
 
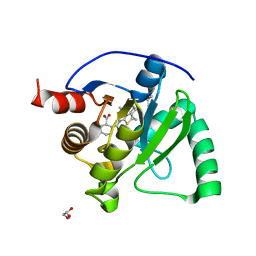 | | SARS-CoV-2 Mac1 in complex with MDOLL-0229 | | Descriptor: | (1R,2R)-2-{[3-(methoxycarbonyl)-4,5,6,7,8,9-hexahydrocycloocta[b]thiophen-2-yl]carbamoyl}cyclohexane-1-carboxylic acid, GLYCEROL, Papain-like protease nsp3 | | Authors: | Wazir, S, Maksimainen, M, Lehtio, L. | | Deposit date: | 2023-08-17 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of 2-Amide-3-methylester Thiophenes that Target SARS-CoV-2 Mac1 and Repress Coronavirus Replication, Validating Mac1 as an Antiviral Target.
J.Med.Chem., 67, 2024
|
|
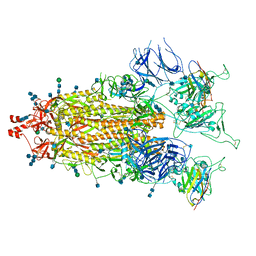
7MY2
 
 | |
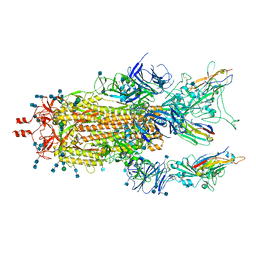
7MY3
 
 | |
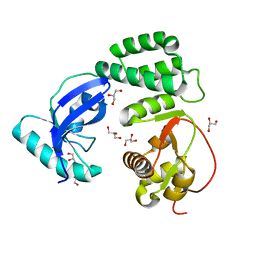
8F2E
 
 | |
8GB6
 
 | |
8GB7
 
 | |
8GB8
 
 | |
8GB5
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody 25F9 | | Descriptor: | 25F9 Heavy chain, 25F9 Light chain, BICINE, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-02-24 | | Release date: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Broadly neutralizing antibodies against sarbecoviruses generated by immunization of macaques with an AS03-adjuvanted COVID-19 vaccine.
Sci Transl Med, 15, 2023
|
|
6WEY
 
 | |
7RB2
 
 | | Cryo-EM structure of SARS-CoV-2 NSP15 NendoU in BIS-Tris pH 6.0 | | Descriptor: | Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Song, Y, Nakamura, A.M, Noske, G.D, Gawriljuk, V.O, Fernandes, R.S, Oliva, G. | | Deposit date: | 2021-07-05 | | Release date: | 2021-07-14 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 51, 2023
|
|
7RB0
 
 | | Cryo-EM structure of SARS-CoV-2 NSP15 NendoU at pH 7.5 | | Descriptor: | Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Song, Y, Nakamura, A.M, Noske, G.D, Gawriljuk, V.O, Fernandes, R.S, Oliva, G. | | Deposit date: | 2021-07-05 | | Release date: | 2021-07-14 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 51, 2023
|
|
7C8T
 
 | | Complex Structure of SARS-CoV-2 3CL Protease with TG-0205221 | | Descriptor: | 3C-like proteinase, N-[(BENZYLOXY)CARBONYL]-O-(TERT-BUTYL)-L-THREONYL-3-CYCLOHEXYL-N-[(1S)-2-HYDROXY-1-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}ETHYL]-L-ALANINAMIDE | | Authors: | Lee, C.C, Wang, A.H.J, Kuo, C.J, Liang, P.H. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Complex Structures and Cellular Activities of the Potent SARS-CoV-2 3CLpro Inhibitors Guiding Drug Discovery Against COVID-19
To Be Published
|
|
7C8R
 
 | | Complex Structure of SARS-CoV-2 3CL Protease with TG-0203770 | | Descriptor: | 3C-like proteinase, ethyl (4R)-4-[[(2S)-4-methyl-2-[[(2S,3R)-3-[(2-methylpropan-2-yl)oxy]-2-(phenylmethoxycarbonylamino)butanoyl]amino]pentanoyl]amino]-5-[(3S)-2-oxidanylidenepyrrolidin-3-yl]pentanoate | | Authors: | Lee, C.C, Wang, A.H.J, Kuo, C.J, Liang, P.H. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Complex Structures and Cellular Activities of the Potent SARS-CoV-2 3CLpro Inhibitors Guiding Drug Discovery Against COVID-19
To Be Published
|
|
7D4F
 
 | | Structure of COVID-19 RNA-dependent RNA polymerase bound to suramin | | Descriptor: | 8-(3-(3-aminobenzamido)-4-methylbenzamido)naphthalene-1,3,5-trisulfonic acid, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Li, Z, Yin, W, Zhou, Z, Yu, X, Xu, H. | | Deposit date: | 2020-09-23 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 RNA polymerase by suramin.
Nat.Struct.Mol.Biol., 28, 2021
|
|
8U1G
 
 | | Prefusion-stabilized SARS-CoV-2 S2 subunit | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S2 | | Authors: | Hsieh, C.-L, McLellan, J.S. | | Deposit date: | 2023-08-31 | | Release date: | 2024-01-24 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Prefusion-stabilized SARS-CoV-2 S2-only antigen provides protection against SARS-CoV-2 challenge.
Nat Commun, 15, 2024
|
|
8EUA
 
 | | Structure of SARS-CoV2 PLpro bound to a covalent inhibitor | | Descriptor: | Papain-like protease nsp3, SULFATE ION, ZINC ION, ... | | Authors: | Mathews, I.I, Pokhrel, S, Wakatsuki, S. | | Deposit date: | 2022-10-18 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Potent and selective covalent inhibition of the papain-like protease from SARS-CoV-2.
Nat Commun, 14, 2023
|
|
7XOG
 
 | | Cryo-EM structure of S glycoprotein encoded by the Covid-19 mRNA vaccine candidate RQ3013 (Postfusion state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,peptide, ... | | Authors: | Wu, Z, Yu, Z, Tan, S, Lu, J, Lu, G, Lin, J. | | Deposit date: | 2022-05-01 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Preclinical evaluation of RQ3013, a broad-spectrum mRNA vaccine against SARS-CoV-2 variants.
Sci Bull (Beijing), 68, 2023
|
|
8DIC
 
 | |
8DIF
 
 | |
8DIB
 
 | |
8DIH
 
 | | Virtual screening for novel SARS-CoV-2 main protease non-covalent and covalent inhibitors | | Descriptor: | (1P,1'R)-1-(isoquinolin-4-yl)-2',3'-dihydrospiro[imidazolidine-4,1'-indene]-2,5-dione, 3C-like proteinase nsp5 | | Authors: | Singh, I, Shoichet, B.K. | | Deposit date: | 2022-06-29 | | Release date: | 2023-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Large library docking for novel SARS-CoV-2 main protease non-covalent and covalent inhibitors.
Protein Sci., 32, 2023
|
|
8DIG
 
 | |
8DII
 
 | |
8DIE
 
 | |
8DID
 
 | |
