8EYL
 
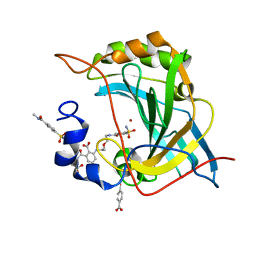 | | Human Carbonic Anhydrase II with Tert-butyl (2-(2-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)ethoxy)ethyl)carbamate | | Descriptor: | 2-[[(2~{R})-1-azanyl-5-oxidanyl-1,5-bis(oxidanylidene)pentan-2-yl]carbamoyl]-6-[2-[2-[(4-sulfamoylphenyl)carbonylamino]ethoxy]ethylamino]benzoic acid, Carbonic anhydrase 2, MERCURIBENZOIC ACID, ... | | Authors: | Kohlbrand, A.J, O'Herin, C.B. | | Deposit date: | 2022-10-27 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Development of Human Carbonic Anhydrase II Heterobifunctional Degraders.
J.Med.Chem., 2023
|
|
7D79
 
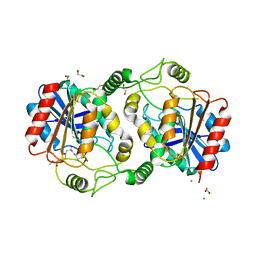 | | The structure of DcsB complex with its substrate analogue | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DltD domain-containing protein, ... | | Authors: | Tang, Y, Zhou, J.H, Wang, G.Q. | | Deposit date: | 2020-10-03 | | Release date: | 2021-01-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.10411429 Å) | | Cite: | A Polyketide Cyclase That Forms Medium-Ring Lactones.
J.Am.Chem.Soc., 143, 2021
|
|
7S7S
 
 | |
1EAD
 
 | |
7A6P
 
 | |
7SBH
 
 | | Crystal structure of the iron superoxide dismutase from Acinetobacter sp. Ver3 | | Descriptor: | FE (III) ION, FLAVIN MONONUCLEOTIDE, Superoxide dismutase | | Authors: | Steimbruch, B.A, Albanesi, D, Repizo, G.D, Lisa, M.N. | | Deposit date: | 2021-09-24 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | The distinctive roles played by the superoxide dismutases of the extremophile Acinetobacter sp. Ver3.
Sci Rep, 12, 2022
|
|
7DS8
 
 | |
7DS3
 
 | |
7DS6
 
 | |
7DS4
 
 | |
7DS2
 
 | |
7DSB
 
 | |
7D85
 
 | |
7V5Y
 
 | |
7V5Z
 
 | |
7DP3
 
 | | Human MCM8 N-terminal domain | | Descriptor: | DNA helicase MCM8, ZINC ION | | Authors: | Li, J, Liu, L, Liu, Y. | | Deposit date: | 2020-12-17 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural study of the N-terminal domain of human MCM8/9 complex.
Structure, 29, 2021
|
|
7VSG
 
 | |
7VSH
 
 | |
7DQK
 
 | | A nicotine MATE transporter, Nicotiana tabacum MATE2 (NtMATE2) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tanaka, Y, Tsukazaki, T, Sasaki, A, Iwaki, S. | | Deposit date: | 2020-12-24 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of a nicotine MATE transporter provide insight into its mechanism of substrate transport.
Febs Lett., 595, 2021
|
|
5F1M
 
 | |
7S8D
 
 | | Structure of DNA-free SgrAI | | Descriptor: | CALCIUM ION, SgraIR restriction enzyme | | Authors: | Horton, N.C. | | Deposit date: | 2021-09-17 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Pretransition state and apo structures of the filament-forming enzyme SgrAI elucidate mechanisms of activation and substrate specificity.
J.Biol.Chem., 298, 2022
|
|
7DDE
 
 | | Cryo-EM structure of the Ape4 and Nbr1 complex | | Descriptor: | Aspartyl aminopeptidase 1,ZZ-type zinc finger-containing protein P35G2.11c,Maltose/maltodextrin-binding periplasmic protein, ZINC ION | | Authors: | Zhang, J, Ye, K. | | Deposit date: | 2020-10-28 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | Molecular and structural mechanisms of ZZ domain-mediated cargo selection by Nbr1.
Embo J., 40, 2021
|
|
7DD9
 
 | | Cryo-EM structure of the Ams1 and Nbr1 complex | | Descriptor: | Alpha-mannosidase,ZZ-type zinc finger-containing protein P35G2.11c,Maltose/maltodextrin-binding periplasmic protein, ZINC ION | | Authors: | Zhang, J, Ye, K. | | Deposit date: | 2020-10-28 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Molecular and structural mechanisms of ZZ domain-mediated cargo selection by Nbr1.
Embo J., 40, 2021
|
|
5EZD
 
 | | Crystal structure of a Hepatocyte growth factor activator inhibitor-1 (HAI-1) fragment covering the PKD-like 'internal' domain and Kunitz domain 1 | | Descriptor: | ACETATE ION, Kunitz-type protease inhibitor 1, POTASSIUM ION | | Authors: | Hong, Z, Andreasen, P.A, Morth, J.P, Jensen, J.K. | | Deposit date: | 2015-11-26 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of a Two-domain Fragment of Hepatocyte Growth Factor Activator Inhibitor-1: FUNCTIONAL INTERACTIONS BETWEEN THE KUNITZ-TYPE INHIBITOR DOMAIN-1 AND THE NEIGHBORING POLYCYSTIC KIDNEY DISEASE-LIKE DOMAIN.
J.Biol.Chem., 291, 2016
|
|
7S7I
 
 | | Crystal structure of Fab in complex with MICA alpha3 domain | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab heavy chain, ... | | Authors: | Lee, P.S, Strop, P. | | Deposit date: | 2021-09-16 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Residue-Level Characterization of Antibody Binding Epitopes Using Carbene Chemical Footprinting.
Anal.Chem., 95, 2023
|
|
