2Y7N
 
 | |
2Y7X
 
 | | The discovery of potent and long-acting oral factor Xa inhibitors with tetrahydroisoquinoline and benzazepine P4 motifs | | Descriptor: | 6-CHLORO-N-[(3S)-1-(5-FLUORO-1,2,3,4-TETRAHYDROISOQUINOLIN-6-YL)-2-OXO-PYRROLIDIN-3-YL]NAPHTHALENE-2-SULFONAMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Watson, N.S, Adams, C, Belton, D, Brown, D, Burns-Kurtis, C.L, Chaudry, L, Chan, C, Convery, M.A, Davies, D.E, Exall, A.M, Harling, J.D, Irving, W.R, Irvine, S, Kleanthous, S, McLay, I.M, Pateman, A.J, Patikis, A.N, Roethka, T.J, Senger, S, Stelman, G.J, Toomey, J.R, West, R.I, Whittaker, C, Zhou, P, Young, R.J. | | Deposit date: | 2011-02-02 | | Release date: | 2011-03-16 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Discovery of Potent and Long-Acting Oral Factor Xa Inhibitors with Tetrahydroisoquinoline and Benzazepine P4 Motifs.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2YOC
 
 | | Crystal structure of PulA from Klebsiella oxytoca | | Descriptor: | CALCIUM ION, PULLULANASE, SULFATE ION | | Authors: | Francetic, O, Mechaly, A.E, Tello-Manigne, D, Buschiazzo, A, Bernarde, C, Nadeau, N, Pugsley, A.P, Alzari, P.M. | | Deposit date: | 2012-10-23 | | Release date: | 2013-11-06 | | Last modified: | 2016-01-20 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural Basis of Pullulanase Membrane Binding and Secretion Revealed by X-Ray Crystallography, Molecular Dynamics and Biochemical Analysis
Structure, 24, 2016
|
|
6WQ0
 
 | | Cryo-EM of the S. solfataricus rod-shaped virus, SSRV1 | | Descriptor: | DNA (301-MER), Structural protein | | Authors: | Wang, F, Baquero, D.P, Beltran, L.C, Prangishvili, D, Krupovic, M, Egelman, E.H. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of filamentous viruses infecting hyperthermophilic archaea explain DNA stabilization in extreme environments.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2YRR
 
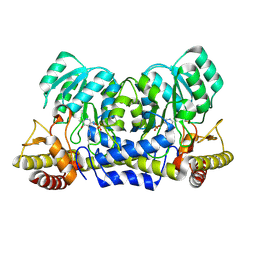 | | hypothetical alanine aminotransferase (TTH0173) from Thermus thermophilus HB8 | | Descriptor: | Aminotransferase, class V, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Miyahara, I, Matsumura, M, Goto, M, Omi, R, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-02 | | Release date: | 2008-04-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | hypothetical alanine aminotransferase (TTH0173) from Thermus thermophilus HB8
To be Published
|
|
6WPR
 
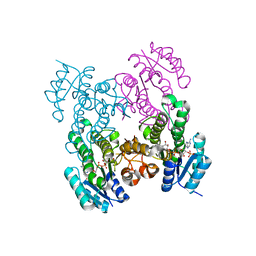 | |
7C2W
 
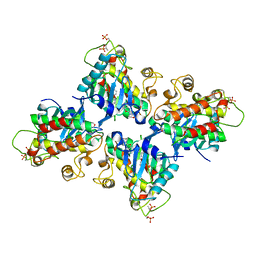 | |
7CL5
 
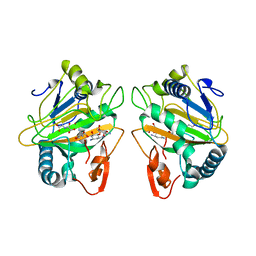 | | The crystal structure of KanJ in complex with kanamycin B and N-oxalylglycine | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-DIAMINO-3-[(3-AMINO-3-DEOXY-ALPHA-D-GLUCOPYRANOSYL)OXY]-2-HYDROXYCYCLOHEXYL 2,6-DIAMINO-2,6-DIDEOXY-ALPHA-D-GLUCOPYRANOSIDE, Kanamycin B dioxygenase, N-OXALYLGLYCINE, ... | | Authors: | Kitayama, Y, Miyanaga, A, Kudo, F, Eguchi, T. | | Deposit date: | 2020-07-20 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Stepwise Post-glycosylation Modification of Sugar Moieties in Kanamycin Biosynthesis.
Chembiochem, 22, 2021
|
|
6WQD
 
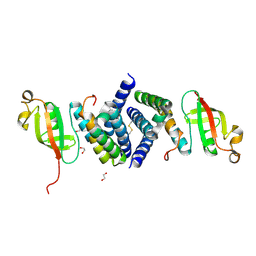 | | The 1.95 A Crystal Structure of the Co-factor Complex of NSP7 and the C-terminal Domain of NSP8 from SARS-CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, Non-structural protein 7, Non-structural protein 8 | | Authors: | Kim, Y, Wilamowski, M, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-28 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Transient and stabilized complexes of Nsp7, Nsp8, and Nsp12 in SARS-CoV-2 replication.
Biophys.J., 120, 2021
|
|
2YJC
 
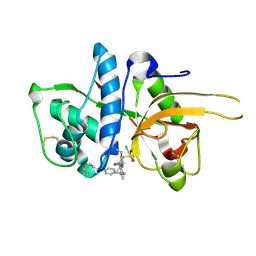 | | CATHEPSIN L WITH A NITRILE INHIBITOR | | Descriptor: | (2S,4R)-1-[1-(4-chlorophenyl)cyclopropyl]carbonyl-4-(2-chlorophenyl)sulfonyl-N-[1-(iminomethyl)cyclopropyl]pyrrolidine-2-carboxamide, CATHEPSIN L1 | | Authors: | Banner, D.W, Benz, J.M, Haap, W. | | Deposit date: | 2011-05-19 | | Release date: | 2011-11-23 | | Last modified: | 2011-11-30 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Halogen Bonding at the Active Sites of Human Cathepsin L and Mek1 Kinase: Efficient Interactions in Different Environments.
Chemmedchem, 6, 2011
|
|
7C6X
 
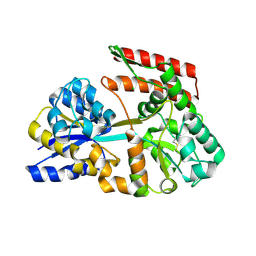 | | Crystal structure of beta-glycosides-binding protein (W41A) of ABC transporter in an open state (Form I) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-22 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
2YJL
 
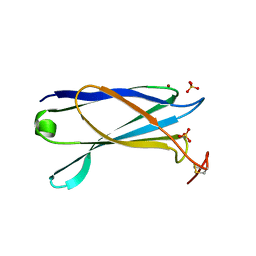 | | Structural characterization of a secretin pilot protein from the type III secretion system (T3SS) of Pseudomonas aeruginosa | | Descriptor: | EXOENZYME S SYNTHESIS PROTEIN B, NICKEL (II) ION, SULFATE ION | | Authors: | Izore, T, Perdu, C, Job, V, Atree, I, Faudry, E, Dessen, A. | | Deposit date: | 2011-05-20 | | Release date: | 2011-08-10 | | Last modified: | 2011-10-12 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural Characterization and Membrane Localization of Exsb from the Type III Secretion System (T3Ss) of Pseudomonas Aeruginosa
J.Mol.Biol., 413, 2011
|
|
2YWI
 
 | |
2YWQ
 
 | | Crystal structure of Thermus thermophilus Protein Y N-terminal domain | | Descriptor: | Ribosomal subunit interface protein | | Authors: | Kawazoe, M, Takemoto, C, Kaminishi, T, Tatsuguchi, A, Saito, Y, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-21 | | Release date: | 2008-04-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structure of Thermus thermophilus Protein Y N-terminal domain
To be Published
|
|
2YX8
 
 | | Crystal structure of the extracellular domain of human RAMP1 | | Descriptor: | Receptor activity-modifying protein 1 | | Authors: | Kusano, S, Kukimoto-Niino, M, Shirouzu, M, Shindo, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-24 | | Release date: | 2008-04-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the human receptor activity-modifying protein 1 extracellular domain.
Protein Sci., 17, 2008
|
|
7CCE
 
 | |
2YZ1
 
 | |
4K1J
 
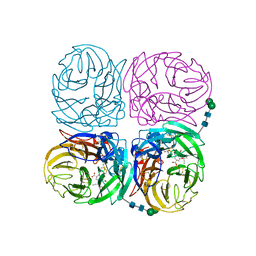 | | Induced opening of influenza virus neuraminidase N2 150-loop suggests an important role in inhibitor binding | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wu, Y, Gao, F, Qi, J.X, Gao, G.F. | | Deposit date: | 2013-04-05 | | Release date: | 2013-06-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Induced opening of influenza virus neuraminidase N2 150-loop suggests an important role in inhibitor binding
Sci Rep, 3, 2013
|
|
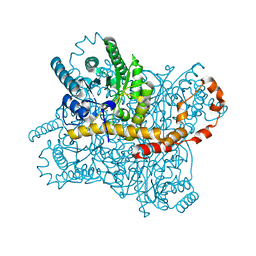
7CK0
 
 | |
6X1U
 
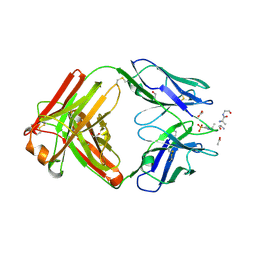 | | Structure of pHis Fab (SC39-4) in complex with pHis mimetic peptide | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ACLYana-3-pTza peptide, ... | | Authors: | Kalagiri, R, Stanfield, R, Wilson, I.A, Hunter, T. | | Deposit date: | 2020-05-19 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | Structural basis for differential recognition of phosphohistidine-containing peptides by 1-pHis and 3-pHis monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2YMW
 
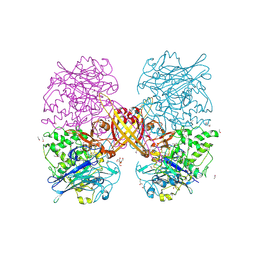 | |
7C5E
 
 | | Crystal structure of Keap1 in complex with fumarate (FUM) | | Descriptor: | ACETATE ION, FUMARIC ACID, Kelch-like ECH-associated protein 1, ... | | Authors: | Padmanabhan, B, Unni, S, Deshmukh, P. | | Deposit date: | 2020-05-19 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights into the multiple binding modes of Dimethyl Fumarate (DMF) and its analogs to the Kelch domain of Keap1.
Febs J., 288, 2021
|
|
7CMD
 
 | | Crystal structure of the SARS-CoV-2 PLpro with GRL0617 | | Descriptor: | 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide, Non-structural protein 3, ZINC ION | | Authors: | Gao, X, Cui, S. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of SARS-CoV-2 papain-like protease.
Acta Pharm Sin B, 11, 2021
|
|
7CNG
 
 | | Structure of CDK5R1 bound FEM1B | | Descriptor: | Protein fem-1 homolog B,Peptide from Cyclin-dependent kinase 5 activator 1, SULFATE ION | | Authors: | Chen, X, Liao, S, Xu, C. | | Deposit date: | 2020-07-31 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
6WZJ
 
 | | LY3041658 Fab bound to CXCL2 | | Descriptor: | C-X-C motif chemokine 2, LY3041658 Fab Heavy Chain, LY3041658 Fab Light Chain | | Authors: | Durbin, J.D, Druzina, Z. | | Deposit date: | 2020-05-14 | | Release date: | 2020-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Discovery and characterization of a neutralizing pan-ELR+CXC chemokine monoclonal antibody.
Mabs, 12
|
|
