8Y1U
 
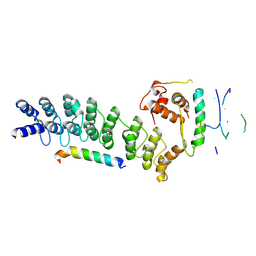 | | Crystal structure of ASB7-Elongin B/C bound to the LZTS1-degron | | Descriptor: | Ankyrin repeat and SOCS box protein 7, Elongin-B, Elongin-C, ... | | Authors: | Dong, C, Yan, X, Zhou, M. | | Deposit date: | 2024-01-25 | | Release date: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Molecular insights into degron recognition by CRL5 ASB7 ubiquitin ligase.
Nat Commun, 15, 2024
|
|
7JP1
 
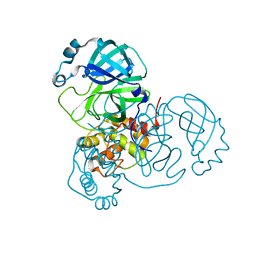 | |
5WE3
 
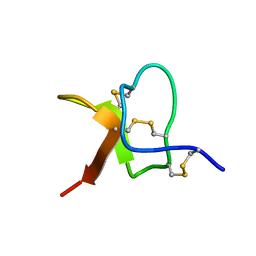 | | Solution NMR structure of PaurTx-3 | | Descriptor: | Beta-theraphotoxin-Ps1a | | Authors: | Agwa, A.J, Schroeder, C.I. | | Deposit date: | 2017-07-06 | | Release date: | 2017-09-13 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Lengths of the C-Terminus and Interconnecting Loops Impact Stability of Spider-Derived Gating Modifier Toxins.
Toxins (Basel), 9, 2017
|
|
9AU5
 
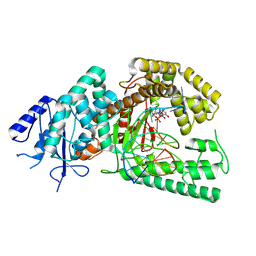 | |
6KKP
 
 | |
5CON
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-015 | | Descriptor: | (3R,3aS,4S,7aS)-3-hydroxyhexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, HIV-1 protease | | Authors: | Yedidi, R.S, Hayashi, H, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2015-07-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | C-5-Modified Tetrahydropyrano-Tetrahydofuran-Derived Protease Inhibitors (PIs) Exert Potent Inhibition of the Replication of HIV-1 Variants Highly Resistant to Various PIs, including Darunavir.
J.Virol., 90, 2015
|
|
5COO
 
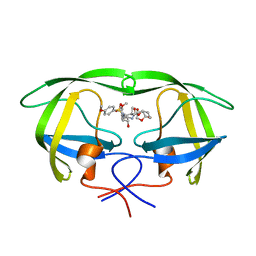 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-085 | | Descriptor: | (3R,3aS,4S,7aS)-3-hydroxyhexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-3-hydroxy-1-(4-methoxyphenyl)-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}butan-2-yl]carbamate, HIV-1 protease | | Authors: | Yedidi, R.S, Hayashi, H, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2015-07-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | C-5-Modified Tetrahydropyrano-Tetrahydofuran-Derived Protease Inhibitors (PIs) Exert Potent Inhibition of the Replication of HIV-1 Variants Highly Resistant to Various PIs, including Darunavir.
J.Virol., 90, 2015
|
|
3GTN
 
 | |
8ASJ
 
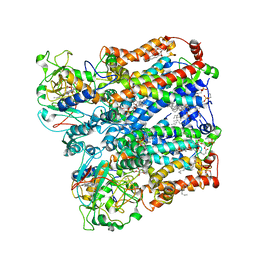 | | Four subunit cytochrome b-c1 complex from Rhodobacter sphaeroides in native nanodiscs - focussed refinement in the b-c conformation | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Cytochrome b, Cytochrome b-c1 subunit IV, ... | | Authors: | Swainsbury, D.J.K, Hawkings, F.R, Martin, E.C, Musial, S, Salisbury, J.H, Jackson, P.J, Farmer, D.A, Johnson, M.P, Siebert, C.A, Hitchcock, A, Hunter, C.N. | | Deposit date: | 2022-08-19 | | Release date: | 2023-03-15 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Cryo-EM structure of the four-subunit Rhodobacter sphaeroides cytochrome bc 1 complex in styrene maleic acid nanodiscs.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6QNO
 
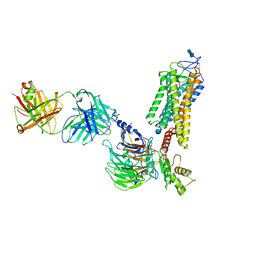 | | Rhodopsin-Gi protein complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab antibody fragment heavy chain, Fab antibody fragment light chain, ... | | Authors: | Tsai, C.-J, Marino, J, Adaixo, R.J, Pamula, F, Muehle, J, Maeda, S, Flock, T, Taylor, N.M.I, Mohammed, I, Matile, H, Dawson, R.J.P, Deupi, X, Stahlberg, H, Schertler, G.F.X. | | Deposit date: | 2019-02-11 | | Release date: | 2019-07-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.38 Å) | | Cite: | Cryo-EM structure of the rhodopsin-G alpha i-beta gamma complex reveals binding of the rhodopsin C-terminal tail to the G beta subunit.
Elife, 8, 2019
|
|
7PD2
 
 | | Crystal structure of the substrate-free radical SAM tyrosine lyase ThiH (2-iminoacetate synthase) from Thermosinus carboxydivorans | | Descriptor: | 5'-DEOXYADENOSINE, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Amara, P, Saragaglia, C, Mouesca, J.-M, Martin, L, Nicolet, Y. | | Deposit date: | 2021-08-04 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | L-tyrosine-bound ThiH structure reveals C-C bond break differences within radical SAM aromatic amino acid lyases.
Nat Commun, 13, 2022
|
|
7PD1
 
 | | Crystal structure of the L-tyrosine-bound radical SAM tyrosine lyase ThiH (2-iminoacetate synthase) from Thermosinus carboxydivorans | | Descriptor: | 5'-DEOXYADENOSINE, BROMIDE ION, GLYCEROL, ... | | Authors: | Amara, P, Saragaglia, C, Mouesca, J.-M, Martin, L, Nicolet, Y. | | Deposit date: | 2021-08-04 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | L-tyrosine-bound ThiH structure reveals C-C bond break differences within radical SAM aromatic amino acid lyases.
Nat Commun, 13, 2022
|
|
2KDK
 
 | |
8V0Y
 
 | |
6RQF
 
 | | 3.6 Angstrom cryo-EM structure of the dimeric cytochrome b6f complex from Spinacia oleracea with natively bound thylakoid lipids and plastoquinone molecules | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Malone, L.A, Qian, P, Mayneord, G.E, Hitchcock, A, Farmer, D, Thompson, R, Swainsbury, D.J.K, Ranson, N, Hunter, C.N, Johnson, M.P. | | Deposit date: | 2019-05-15 | | Release date: | 2019-11-13 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Cryo-EM structure of the spinach cytochrome b6f complex at 3.6 angstrom resolution.
Nature, 575, 2019
|
|
8USU
 
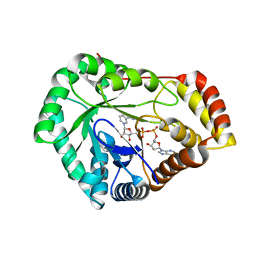 | | Crystal Structure of L-galactose 1-dehydrogenase of Myrciaria dubia in complex with NAD | | Descriptor: | L-galactose dehydrogenase isoform X1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Santillan, J.A.V, Cabrejos, D.A.L, Pereira, H.M, Gomez, J.C.C, Garratt, R.C. | | Deposit date: | 2023-10-30 | | Release date: | 2024-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Structural insights into the Smirnoff-Wheeler pathway for vitamin C production in the Amazon fruit camu-camu.
J.Exp.Bot., 75, 2024
|
|
7ST8
 
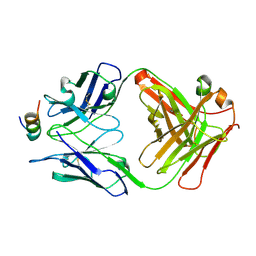 | | Crystal structure of 7H2.2 Fab in complex with SAS1B C-terminal region | | Descriptor: | 7H2.2 Fab Heavy Chain, 7H2.2 Fab Light Chain, Astacin-like metalloendopeptidase | | Authors: | Legg, M.S.G, Evans, S.V. | | Deposit date: | 2021-11-12 | | Release date: | 2022-05-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Monoclonal antibody 7H2.2 binds the C-terminus of the cancer-oocyte antigen SAS1B through the hydrophilic face of a conserved amphipathic helix corresponding to one of only two regions predicted to be ordered
Acta Crystallogr.,Sect.D, 78, 2022
|
|
6XAR
 
 | |
8DVX
 
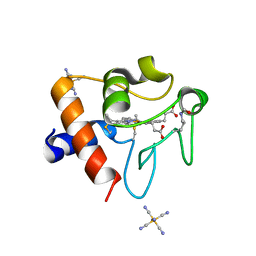 | | Structure of acetylated Pig somatic Cytochrome c (Aly39) at 1.5A | | Descriptor: | Cytochrome c, HEME C, HEXACYANOFERRATE(3-) | | Authors: | Edwards, B.F.P, Huettemann, M, Vaishnav, A, Brunzelle, J, Morse, P, Wan, J. | | Deposit date: | 2022-07-30 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Cytochrome c lysine acetylation regulates cellular respiration and cell death in ischemic skeletal muscle.
Nat Commun, 14, 2023
|
|
7SML
 
 | | Crystal Structure of L-GALACTONO-1,4-LACTONE DEHYDROGENASE de Myrciaria dubia | | Descriptor: | L-GALACTONO-1,4-LACTONE DEHYDROGENASE | | Authors: | Santillan, J.A.V, Cabrejos, D.A.L, Pereira, H.M, Gomez, J.C.C, Garratt, R.C. | | Deposit date: | 2021-10-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the Smirnoff-Wheeler pathway for vitamin C production in the Amazon fruit Camu-Camu.
J.Exp.Bot., 2024
|
|
5HTI
 
 | | Crystal structure of c-Met kinase domain in complex with LXM108 | | Descriptor: | Hepatocyte growth factor receptor, N-[3-fluoro-4-({7-[2-(morpholin-4-yl)ethoxy]-1,6-naphthyridin-4-yl}oxy)phenyl]-N'-(4-fluorophenyl)cyclopropane-1,1-dicarboxamide | | Authors: | Liu, Q.F, Xu, Y.C. | | Deposit date: | 2016-01-26 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of c-Met kinase domain in complex with LXM108
to be published
|
|
6Y4T
 
 | | Crystal structure of p38 in complex with SR63. | | Descriptor: | 1,2-ETHANEDIOL, 5-azanyl-~{N}-[[4-[[(2~{S})-1-[[(2~{S})-butan-2-yl]amino]-4-cyclohexyl-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Chaikuad, A, Roehm, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-23 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Selective targeting of the alpha C and DFG-out pocket in p38 MAPK.
Eur.J.Med.Chem., 208, 2020
|
|
7B42
 
 | | Crystal structure of c-MET bound by compound 8 | | Descriptor: | 3-[(3-fluorophenyl)methyl]-5-(1-piperidin-4-ylpyrazol-4-yl)-1~{H}-pyrrolo[2,3-b]pyridine, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2020-12-02 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Targeting the Folded P-Loop Conformation of c-MET.
Acs Med.Chem.Lett., 12, 2021
|
|
6Y4U
 
 | | Crystal structure of p38 in complex with SR65 | | Descriptor: | 1,2-ETHANEDIOL, 5-azanyl-~{N}-[[4-[[(2~{S})-4-cyclohexyl-1-oxidanylidene-1-(pentan-3-ylamino)butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Chaikuad, A, Roehm, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-23 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Selective targeting of the alpha C and DFG-out pocket in p38 MAPK.
Eur.J.Med.Chem., 208, 2020
|
|
7B40
 
 | | Crystal structure of c-MET bound by compound 6 | | Descriptor: | 3-(phenylmethyl)-5-(1-piperidin-4-ylpyrazol-4-yl)-1~{H}-pyrrolo[2,3-b]pyridine, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2020-12-01 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Basis for Targeting the Folded P-Loop Conformation of c-MET.
Acs Med.Chem.Lett., 12, 2021
|
|
