5DF1
 
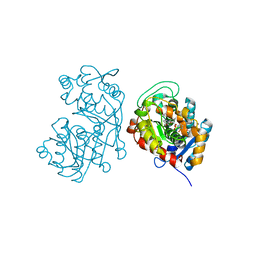 | | Iridoid synthase from Catharanthus roseus - ternary complex with NADP+ and geranic acid | | Descriptor: | (2E)-3,7-dimethylocta-2,6-dienoic acid, 1,2-ETHANEDIOL, IMIDAZOLE, ... | | Authors: | Caputi, L, Kries, H, Stevenson, C.E.M, Kamileen, M.O, Sherden, N.H, Geu-Flores, F, Lawson, D.M, O'Connor, S.E. | | Deposit date: | 2015-08-26 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural determinants of reductive terpene cyclization in iridoid biosynthesis.
Nat.Chem.Biol., 12, 2016
|
|
1GL3
 
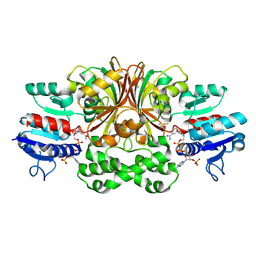 | | ASPARTATE BETA-SEMIALDEHYDE DEHYDROGENASE IN COMPLEX WITH NADP AND SUBSTRATE ANALOGUE S-METHYL CYSTEINE SULFOXIDE | | Descriptor: | ASPARTATE-SEMIALDEHYDE DEHYDROGENASE, CYSTEINE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hadfield, A.T, Kryger, G, Ouyang, J, Ringe, D, Petsko, G.A, Viola, R.E. | | Deposit date: | 2001-08-23 | | Release date: | 2001-11-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Active Site Analysis of the Potential Antimicrobial Target Aspartate Semialdehyde Dehydrogenase.
Biochemistry, 40, 2001
|
|
6ZLF
 
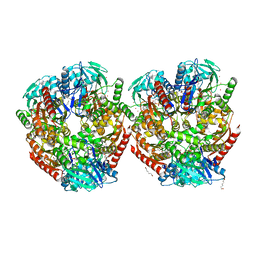 | | Aerobic crystal structure of F420H2-Oxidase from Methanothermococcus thermolithotrophicus at 1.8A resolution under 125 bars of krypton | | Descriptor: | CHLORIDE ION, Coenzyme F420H2 oxidase (FprA), FLAVIN MONONUCLEOTIDE, ... | | Authors: | Engilberge, S, Wagner, T, Carpentier, P, Girard, E, Shima, S. | | Deposit date: | 2020-06-30 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Krypton-derivatization highlights O 2 -channeling in a four-electron reducing oxidase.
Chem.Commun.(Camb.), 56, 2020
|
|
6ZCV
 
 | |
1XJA
 
 | |
6ZHJ
 
 | | 3D electron diffraction structure of thermolysin from Bacillus thermoproteolyticus | | Descriptor: | CALCIUM ION, Thermolysin, ZINC ION | | Authors: | Blum, T, Housset, D, Clabbers, M.T.B, van Genderen, E, Schoehn, G, Ling, W.L, Abrahams, J.P. | | Deposit date: | 2020-06-23 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.26 Å) | | Cite: | Statistically correcting dynamical electron scattering improves the refinement of protein nanocrystals, including charge refinement of coordinated metals.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
3P70
 
 | | Structural basis of thrombin-mediated factor V activation: essential role of the hirudin-like sequence Glu666-Glu672 for processing at the heavy chain-B domain junction | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BENZAMIDINE, ... | | Authors: | Corral-Rodriguez, M.A, Bock, P.E, Hernandez-Carvajal, E, Gutierrez-Gallego, R, Fuentes-Prior, P. | | Deposit date: | 2010-10-11 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis of thrombin-mediated factor V activation: the Glu666-Glu672 sequence is critical for processing at the heavy chain-B domain junction.
Blood, 117, 2011
|
|
6W8U
 
 | | Cryo-EM of the Pyrobaculum arsenaticum pilus | | Descriptor: | pilin | | Authors: | Wang, F, Baquero, D.P, Su, Z, Beltran, L.C, Prangishvili, D, Krupovic, M, Egelman, E.H. | | Deposit date: | 2020-03-21 | | Release date: | 2020-07-08 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The structures of two archaeal type IV pili illuminate evolutionary relationships.
Nat Commun, 11, 2020
|
|
6ZK8
 
 | | Native crystal structure of anaerobic F420H2-Oxidase from Methanothermococcus thermolithotrophicus at 1.8A resolution | | Descriptor: | Coenzyme F420H2 oxidase (FprA), DI(HYDROXYETHYL)ETHER, FE (III) ION, ... | | Authors: | Engilberge, S, Wagner, T, Carpentier, P, Girard, E, Shima, S. | | Deposit date: | 2020-06-30 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Krypton-derivatization highlights O 2 -channeling in a four-electron reducing oxidase.
Chem.Commun.(Camb.), 56, 2020
|
|
1GL4
 
 | | Nidogen-1 G2/Perlecan IG3 Complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BASEMENT MEMBRANE-SPECIFIC HEPARAN SULFATE PROTEOGLYCAN CORE PROTEIN, NIDOGEN-1, ... | | Authors: | Kvansakul, M, Hopf, M, Ries, A, Timpl, R, Hohenester, E. | | Deposit date: | 2001-08-23 | | Release date: | 2001-11-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the High-Affinity Interaction of Nidogen-1 with Immunoglobulin-Like Domain 3 of Perlecan
Embo J., 20, 2001
|
|
1KHQ
 
 | | ORTHORHOMBIC FORM OF PAPAIN/ZLFG-DAM COVALENT COMPLEX | | Descriptor: | Papain, peptidic inhibitor | | Authors: | Janowski, R, Kozak, M, Jankowska, E, Grzonka, Z, Jaskolski, M. | | Deposit date: | 2001-11-30 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Two polymorphs of a covalent complex between papain and a diazomethylketone inhibitor
J.Pept.Res., 64, 2004
|
|
6Z84
 
 | | CK2 alpha bound to chemical probe SGC-CK2-1 derivative | | Descriptor: | Casein kinase II subunit alpha, SULFATE ION, ~{N}-[1-[3-cyano-7-(cyclopropylamino)pyrazolo[1,5-a]pyrimidin-5-yl]indol-6-yl]ethanamide | | Authors: | Kraemer, A, Wells, C, Drewry, D.H, Pickett, J.E, Axtman, A.D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-02 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Development of a potent and selective chemical probe for the pleiotropic kinase CK2.
Cell Chem Biol, 28, 2021
|
|
1GCB
 
 | | GAL6, YEAST BLEOMYCIN HYDROLASE DNA-BINDING PROTEASE (THIOL) | | Descriptor: | GAL6 HG (EMTS) DERIVATIVE, GLYCEROL, MERCURY (II) ION, ... | | Authors: | Joshua-Tor, L, Xu, H.E, Johnston, S.A, Rees, D.C. | | Deposit date: | 1995-07-18 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a conserved protease that binds DNA: the bleomycin hydrolase, Gal6.
Science, 269, 1995
|
|
7UVL
 
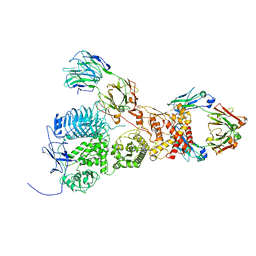 | | IgA1 Protease with IgA1 substrate | | Descriptor: | Immunoglobulin alpha-1 heavy chain, Immunoglobulin alpha-1 heavy constant, Immunoglobulin alpha-1 light chain, ... | | Authors: | Eisenmesser, Z.E, Zheng, H. | | Deposit date: | 2022-05-02 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | A substrate-induced gating mechanism is conserved among Gram-positive IgA1 metalloproteases.
Commun Biol, 5, 2022
|
|
5IH9
 
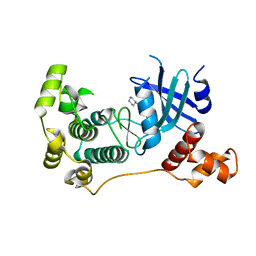 | | MELK in complex with NVS-MELK8A | | Descriptor: | 1-methyl-4-[4-(4-{3-[(piperidin-4-yl)methoxy]pyridin-4-yl}-1H-pyrazol-1-yl)phenyl]piperazine, Maternal embryonic leucine zipper kinase | | Authors: | Sprague, E.R, Puleo, D.E. | | Deposit date: | 2016-02-29 | | Release date: | 2016-06-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Toward the Validation of Maternal Embryonic Leucine Zipper Kinase: Discovery, Optimization of Highly Potent and Selective Inhibitors, and Preliminary Biology Insight.
J.Med.Chem., 59, 2016
|
|
6W6N
 
 | |
1KHP
 
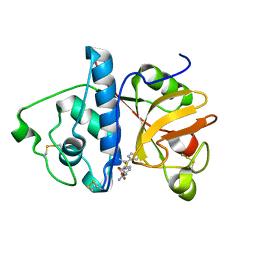 | | Monoclinic form of papain/ZLFG-DAM covalent complex | | Descriptor: | Papain, peptidic inhibitor | | Authors: | Janowski, R, Kozak, M, Jankowska, E, Grzonka, Z, Jaskolski, M. | | Deposit date: | 2001-11-30 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two polymorphs of a covalent complex between papain and a diazomethylketone inhibitor
J.Pept.Res., 64, 2004
|
|
4MCM
 
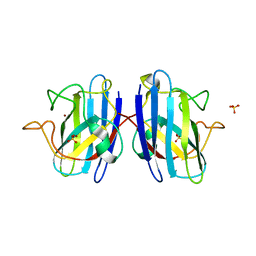 | | Human SOD1 C57S Mutant, As-isolated | | Descriptor: | SULFATE ION, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Sea, K, Sohn, S.H, Durazo, A, Sheng, Y, Shaw, B, Cao, X, Taylor, A.B, Whitson, L.J, Holloway, S.P, Hart, P.J, Cabelli, D.E, Gralla, E.B, Valentine, J.S. | | Deposit date: | 2013-08-21 | | Release date: | 2014-08-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insights into the role of the unusual disulfide bond in copper-zinc superoxide dismutase.
J.Biol.Chem., 290, 2015
|
|
5IIK
 
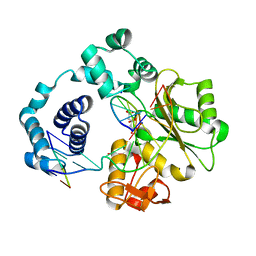 | | Crystal structure of the post-catalytic nick complex of DNA polymerase lambda with a templating 8-oxo-dG and incorporated dC | | Descriptor: | CITRIC ACID, DNA (5'-D(*CP*AP*GP*TP*AP*CP*T)-3'), DNA (5'-D(*CP*GP*GP*CP*AP*(8OG)P*TP*AP*CP*TP*G)-3'), ... | | Authors: | Burak, M.J, Guja, K.E, Garcia-Diaz, M. | | Deposit date: | 2016-03-01 | | Release date: | 2016-08-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.982 Å) | | Cite: | A fidelity mechanism in DNA polymerase lambda promotes error-free bypass of 8-oxo-dG.
Embo J., 35, 2016
|
|
6ZHO
 
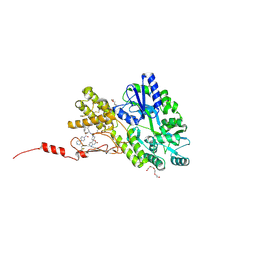 | | Crystal structure of a CGRP receptor ectodomain heterodimer with bound high affinity inhibitor | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Receptor activity-modifying protein 1,Calcitonin gene-related peptide type 1 receptor, TETRAETHYLENE GLYCOL, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Southall, S.M. | | Deposit date: | 2020-06-23 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Based Drug Discovery ofN-((R)-3-(7-Methyl-1H-indazol-5-yl)-1-oxo-1-(((S)-1-oxo-3-(piperidin-4-yl)-1-(4-(pyridin-4-yl)piperazin-1-yl)propan-2-yl)amino)propan-2-yl)-2'-oxo-1',2'-dihydrospiro[piperidine-4,4'-pyrido[2,3-d][1,3]oxazine]-1-carboxamide (HTL22562): A Calcitonin Gene-Related Peptide Receptor Antagonist for Acute Treatment of Migraine.
J.Med.Chem., 63, 2020
|
|
7ZSD
 
 | | cryo-EM structure of omicron spike in complex with de novo designed binder, local | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, de novo designed binder | | Authors: | Pablo, G, Sarah, W, Alexandra, V.H, Anthony, M, Andreas, S, Zander, H, Dongchun, N, Shuguang, T, Freyr, S, Casper, G, Priscilla, T, Alexandra, T, Stephane, R, Sandrine, G, Jane, M, Aaron, P, Zepeng, X, Yan, C, Pu, H, George, G, Elisa, O, Beat, F, Didier, T, Henning, S, Michael, B, Bruno, E.C. | | Deposit date: | 2022-05-06 | | Release date: | 2023-03-01 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
6YSK
 
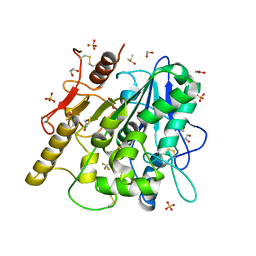 | | 1-phenylpyrroles and 1-enylpyrrolidines as inhibitors of Notum | | Descriptor: | (3~{S})-1-[4-chloranyl-3-(trifluoromethyl)phenyl]pyrrolidine-3-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2020-04-22 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
2UWE
 
 | | Large CDR3a loop alteration as a function of MHC mutation | | Descriptor: | AHIII TCR ALPHA CHAIN, AHIII TCR BETA CHAIN, BETA-2-MICROGLOBULIN, ... | | Authors: | Miller, P.J, Pazy, Y, Conti, B, Riddle, D, Biddison, W.E, Appella, E, Collins, E.J. | | Deposit date: | 2007-03-20 | | Release date: | 2007-09-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Single Mhc Mutation Eliminates Enthalpy Associated with T Cell Receptor Binding.
J.Mol.Biol., 373, 2007
|
|
5ELV
 
 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J-L504-N775) in complex with glutamate and BPAM-521 at 1.92 A resolution | | Descriptor: | 4-Cyclopropyl-3,4-dihydro-7-hydroxy-2H-1,2,4-benzothiadiazine 1,1-dioxide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Krintel, C, Juknaite, L, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2015-11-05 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Enthalpy-Entropy Compensation in the Binding of Modulators at Ionotropic Glutamate Receptor GluA2.
Biophys.J., 110, 2016
|
|
7R4N
 
 | | Structure of human hydroxyacid oxidase 1 bound with 5-bromo-N-methyl-1H-indazole-3-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 5-bromanyl-N-methyl-1H-indazole-3-carboxamide, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Mackinnon, S, Bezerra, G.A, Krojer, T, Bradley, A.R, Talon, R, Brandeo-Neto, J, Douangamath, A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Oppermann, U, Brennan, P.E, Yue, W.W. | | Deposit date: | 2022-02-08 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of human hydroxyacid oxidase 1 bound with 5-bromo-N-methyl-1H-indazole-3-carboxamide
To Be Published
|
|
