6FXC
 
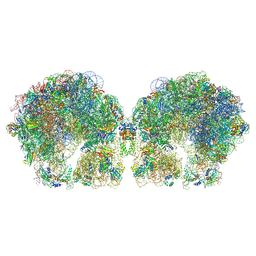 | | The cryo-EM structure of hibernating 100S ribosome dimer from pathogenic Staphylococcus aureus | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Matzov, D, Aibara, S, Zimmerman, E, Bashan, A, Kidmose, R, Amunts, A, Yonath, A. | | Deposit date: | 2018-03-08 | | Release date: | 2018-03-21 | | Last modified: | 2018-11-21 | | Method: | ELECTRON MICROSCOPY (6.76 Å) | | Cite: | The cryo-EM structure of hibernating 100S ribosome dimer from pathogenic Staphylococcus aureus.
Nat Commun, 8, 2017
|
|
6G0R
 
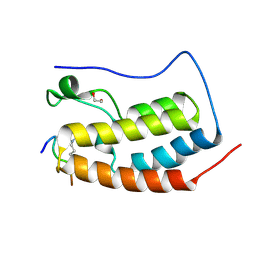 | | Crystal Structure of the first bromodomain of human BRD4 in complex with an acetylated POLR2A peptide (K775ac/K778ac) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, DNA-directed RNA polymerase II subunit RPB1 | | Authors: | Filippakopoulos, P, Picaud, S, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C. | | Deposit date: | 2018-03-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol. Cell, 73, 2019
|
|
5WEA
 
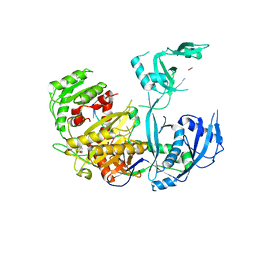 | | Human Argonaute2 Helix-7 Mutant | | Descriptor: | PHENOL, Protein argonaute-2, RNA (5'-R(P*AP*AP*AP*AP*AP*AP*AP*U)-3') | | Authors: | Schirle, N.T, Klum, S.M, MacRae, I.J. | | Deposit date: | 2017-07-08 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Helix-7 in Argonaute2 shapes the microRNA seed region for rapid target recognition.
EMBO J., 37, 2018
|
|
2FO1
 
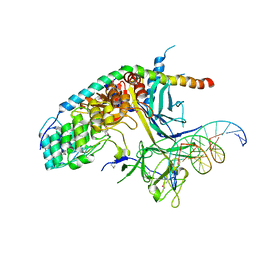 | | Crystal Structure of the CSL-Notch-Mastermind ternary complex bound to DNA | | Descriptor: | 5'-D(*AP*AP*TP*CP*TP*TP*TP*CP*CP*CP*AP*CP*AP*GP*T)-3', 5'-D(*TP*TP*AP*CP*TP*GP*TP*GP*GP*GP*AP*AP*AP*GP*A)-3', Lin-12 and glp-1 phenotype protein 1, ... | | Authors: | Wilson, J.J, Kovall, R.A. | | Deposit date: | 2006-01-12 | | Release date: | 2006-03-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Crystal structure of the CSL-Notch-Mastermind ternary complex bound to DNA.
Cell(Cambridge,Mass.), 124, 2006
|
|
2DIW
 
 | | Solution structure of the RPR domain of Putative RNA-binding protein 16 | | Descriptor: | Putative RNA-binding protein 16 | | Authors: | Dang, W, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-03-30 | | Release date: | 2006-09-30 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RPR domain of Putative RNA-binding protein 16
To be Published
|
|
7EH0
 
 | |
4TUS
 
 | |
3LP3
 
 | | p15 HIV RNaseH domain with inhibitor MK3 | | Descriptor: | 3-[4-(diethylamino)phenoxy]-6-(ethoxycarbonyl)-5,8-dihydroxy-7-oxo-7,8-dihydro-1,8-naphthyridin-1-ium, MANGANESE (II) ION, p15 | | Authors: | Yan, Y, Munshi, S.K, Prasad, G.S, Su, H.P. | | Deposit date: | 2010-02-04 | | Release date: | 2010-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the inhibition of RNase H activity of HIV-1 reverse transcriptase by RNase H active site-directed inhibitors.
J.Virol., 84, 2010
|
|
6PZQ
 
 | |
5TWQ
 
 | | Post-catalytic nicked complex of human Polymerase Mu with newly incorporated UTP | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*CP*GP*GP*CP*AP*TP*AP*CP*G)-3'), ... | | Authors: | Moon, A.F, Pryor, J.M, Ramsden, D.A, Kunkel, T.A, Bebenek, K, Pedersen, L.C. | | Deposit date: | 2016-11-14 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural accommodation of ribonucleotide incorporation by the DNA repair enzyme polymerase Mu.
Nucleic Acids Res., 45, 2017
|
|
3QIP
 
 | | Structure of HIV-1 reverse transcriptase in complex with an RNase H inhibitor and nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, 5,6-dihydroxy-2-[(2-phenyl-1H-indol-3-yl)methyl]pyrimidine-4-carboxylic acid, CHLORIDE ION, ... | | Authors: | Lansdon, E.B, Kirschberg, T.A. | | Deposit date: | 2011-01-27 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.0926 Å) | | Cite: | Structural and Binding Analysis of Pyrimidinol Carboxylic Acid and N-Hydroxy Quinazolinedione HIV-1 RNase H Inhibitors.
Antimicrob.Agents Chemother., 55, 2011
|
|
7V04
 
 | | The N-terminal domain of PA endonuclease from the influenza H1N1 viral polymerase in complex with 6-bromo-3-hydroxy-4-oxo-1,4-dihydropyridine-2-carboxamide | | Descriptor: | 6-bromo-3-hydroxy-4-oxo-1,4-dihydropyridine-2-carboxamide, MANGANESE (II) ION, Polymerase acidic protein | | Authors: | Kohlbrand, A.J, Stokes, R.W, Karges, J, Seo, H, Sankaran, B, Cohen, S.M. | | Deposit date: | 2022-05-09 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Carboxylic Acid Isostere Derivatives of Hydroxypyridinones as Core Scaffolds for Influenza Endonuclease Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
4ZN1
 
 | | Crystal Structure of MjSpt4:Spt5 complex conformation A | | Descriptor: | Transcription elongation factor Spt4, Transcription elongation factor Spt5, ZINC ION | | Authors: | Guo, G.R, Zhou, H.H, Gao, Y.X, Zhu, Z.L, Niu, L.W, Teng, M.K. | | Deposit date: | 2015-05-04 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biochemical insights into the DNA-binding mode of MjSpt4p:Spt5 complex at the exit tunnel of RNAPII
J.Struct.Biol., 192, 2015
|
|
3IG1
 
 | | HIV-1 Reverse Transcriptase with the Inhibitor beta-Thujaplicinol Bound at the RNase H Active Site | | Descriptor: | 2,7-dihydroxy-4-(propan-2-yl)cyclohepta-2,4,6-trien-1-one, HIV-1 Reverse Transcriptase p51 subunit, HIV-1 Reverse Transcriptase p66 subunit, ... | | Authors: | Himmel, D.M, Maegley, K.A, Pauly, T.A, Arnold, E. | | Deposit date: | 2009-07-27 | | Release date: | 2010-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of HIV-1 reverse transcriptase with the inhibitor beta-Thujaplicinol bound at the RNase H active site.
Structure, 17, 2009
|
|
4ZN3
 
 | | Crystal Structure of MjSpt4:Spt5 complex conformation B | | Descriptor: | FE (III) ION, GLYCEROL, Transcription elongation factor Spt4, ... | | Authors: | Guo, G.R, Zhou, H.H, Gao, Y.X, Zhu, Z.L, Niu, L.W, Teng, M.K. | | Deposit date: | 2015-05-04 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and biochemical insights into the DNA-binding mode of MjSpt4p:Spt5 complex at the exit tunnel of RNAPII
J.Struct.Biol., 192, 2015
|
|
3T72
 
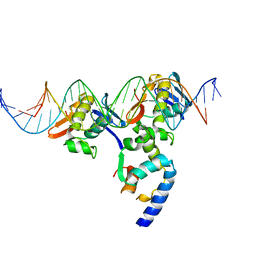 | | PhoB(E)-Sigma70(4)-(RNAP-Betha-flap-tip-helix)-DNA Transcription Activation Sub-Complex | | Descriptor: | PHO BOX DNA (STRAND 1), PHO BOX DNA (STRAND 2), Phosphate regulon transcriptional regulatory protein phoB, ... | | Authors: | Blanco, A.G, Canals, A, Bernues, J, Sola, M, Coll, M. | | Deposit date: | 2011-07-29 | | Release date: | 2011-09-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (4.33 Å) | | Cite: | The structure of a transcription activation subcomplex reveals how sigma (70) is recruited to PhoB promoters.
Embo J., 30, 2011
|
|
4GK0
 
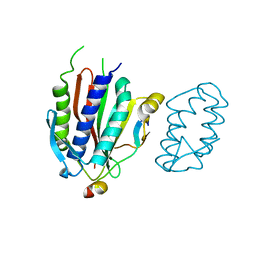 | | Crystal structure of human Rev3-Rev7-Rev1 complex | | Descriptor: | DNA polymerase zeta catalytic subunit, DNA repair protein REV1, Mitotic spindle assembly checkpoint protein MAD2B | | Authors: | Tao, J, Min, X, Wei, X. | | Deposit date: | 2012-08-10 | | Release date: | 2013-03-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the assembly of human translesion polymerase complexes
Protein Cell, 3, 2012
|
|
4GK5
 
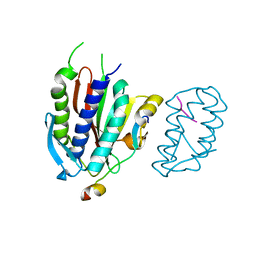 | | Crystal structure of human Rev3-Rev7-Rev1-Polkappa complex | | Descriptor: | DNA polymerase kappa, DNA polymerase zeta catalytic subunit, DNA repair protein REV1, ... | | Authors: | Tao, J, Min, X, Wei, X. | | Deposit date: | 2012-08-10 | | Release date: | 2013-03-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Structural insights into the assembly of human translesion polymerase complexes
Protein Cell, 3, 2012
|
|
5AOR
 
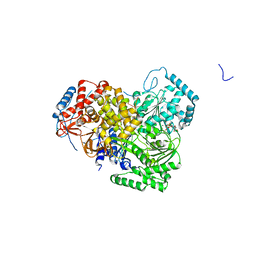 | | Structure of MLE RNA ADP AlF4 complex | | Descriptor: | 5'-R(*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP)-3', ADENOSINE-5'-DIPHOSPHATE, DOSAGE COMPENSATION REGULATOR, ... | | Authors: | Prabu, J.R, Conti, E. | | Deposit date: | 2015-09-11 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure of the RNA Helicase Mle Reveals the Molecular Mechanisms for Uridine Specificity and RNA-ATP Coupling.
Mol.Cell, 60, 2015
|
|
1LV5
 
 | | Crystal Structure of the Closed Conformation of Bacillus DNA Polymerase I Fragment Bound to DNA and dCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*AP*CP*GP*TP*CP*GP*CP*TP*GP*AP*TP*CP*CP*G)-3', 5'-D(*GP*GP*AP*TP*CP*AP*GP*CP*GP*A)-3', ... | | Authors: | Johnson, S.J, Taylor, J.S, Beese, L.S. | | Deposit date: | 2002-05-24 | | Release date: | 2003-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Processive DNA synthesis observed in a polymerase crystal suggests a
mechanism for the prevention of frameshift mutations
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1O0B
 
 | |
7QDZ
 
 | | 80S-bound human SKI complex in the closed state | | Descriptor: | Helicase SKI2W, RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3'), Tetratricopeptide repeat protein 37, ... | | Authors: | Koegel, A, Keidel, A, Bonneau, F, Schaefer, I.B, Conti, E. | | Deposit date: | 2021-12-01 | | Release date: | 2022-02-09 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The human SKI complex regulates channeling of ribosome-bound RNA to the exosome via an intrinsic gatekeeping mechanism.
Mol.Cell, 82, 2022
|
|
1O0C
 
 | |
8ICU
 
 | |
8ICV
 
 | |
