1J1C
 
 | | Binary complex structure of human tau protein kinase I with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glycogen synthase kinase-3 beta, MAGNESIUM ION | | Authors: | Aoki, M, Yokota, T, Sugiura, I, Sasaki, C, Hasegawa, T, Okumura, C, Kohno, T, Sugio, S, Matsuzaki, T. | | Deposit date: | 2002-12-03 | | Release date: | 2003-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insight into nucleotide recognition in tau-protein kinase I/glycogen synthase kinase 3 beta.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1J14
 
 | | BENZAMIDINE IN COMPLEX WITH RAT TRYPSIN MUTANT X99RT | | Descriptor: | BENZAMIDINE, CALCIUM ION, SULFATE ION, ... | | Authors: | Stubbs, M.T. | | Deposit date: | 2002-11-30 | | Release date: | 2002-12-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Reconstructing the Binding Site of Factor Xa in Trypsin Reveals Ligand-induced Structural Plasticity
J.MOL.BIOL., 325, 2003
|
|
3LGV
 
 | | H198P mutant of the DegS-deltaPDZ protease | | Descriptor: | Protease degS | | Authors: | Sohn, J, Grant, R.A, Sauer, R.T. | | Deposit date: | 2010-01-21 | | Release date: | 2010-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.734 Å) | | Cite: | Allostery is an intrinsic property of the protease domain of DegS: implications for enzyme function and evolution.
J.Biol.Chem., 285, 2010
|
|
1J73
 
 | | Crystal structure of an unstable insulin analog with native activity. | | Descriptor: | ZINC ION, insulin a, insulin b | | Authors: | Wan, Z, Zhao, M, Nakagawa, S, Jia, W, Weiss, M.A. | | Deposit date: | 2001-05-15 | | Release date: | 2001-05-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Non-standard insulin design: structure-activity relationships at the periphery of the insulin receptor.
J.Mol.Biol., 315, 2002
|
|
1J82
 
 | | Osmolyte Stabilization of RNase | | Descriptor: | RIBONUCLEASE PANCREATIC, SULFATE ION | | Authors: | Ratnaparkhi, G.S, Varadarajan, R. | | Deposit date: | 2001-05-19 | | Release date: | 2001-06-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Osmolytes stabilize ribonuclease S by stabilizing its fragments S protein and S peptide to compact folding-competent states.
J.Biol.Chem., 276, 2001
|
|
1J8A
 
 | |
1J8K
 
 | | NMR STRUCTURE OF THE FIBRONECTIN EDA DOMAIN, NMR, 20 STRUCTURES | | Descriptor: | FIBRONECTIN | | Authors: | Niimi, T, Osawa, M, Yamaji, N, Yasunaga, K, Sakashita, H, Mase, T, Tanaka, A, Fujita, S. | | Deposit date: | 2001-05-22 | | Release date: | 2002-02-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of human fibronectin EDA.
J.Biomol.NMR, 21, 2001
|
|
3LLH
 
 | | Crystal structure of the first dsRBD of TAR RNA-binding protein 2 | | Descriptor: | MALONATE ION, RISC-loading complex subunit TARBP2 | | Authors: | Yamashita, S, Kawazoe, M, Takemoto, C, Sekine, S, Wakiyama, M, Yokoyama, S. | | Deposit date: | 2010-01-29 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | The structures of dsRBDs of human TRBP
To be Published
|
|
1IFC
 
 | |
1IJB
 
 | | The von Willebrand Factor mutant (I546V) A1 domain | | Descriptor: | von Willebrand factor | | Authors: | Fukuda, K, Doggett, T.A, Bankston, L.A, Cruz, M.A, Diacovo, T.G, Liddington, R.C. | | Deposit date: | 2001-04-25 | | Release date: | 2002-07-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of von Willebrand factor activation by the snake toxin botrocetin.
Structure, 10, 2002
|
|
3LNK
 
 | | Structure of BACE bound to SCH743813 | | Descriptor: | Beta-secretase 1, L(+)-TARTARIC ACID, N'-{(1S,2S)-1-(3,5-difluorobenzyl)-2-hydroxy-2-[(2R)-4-(phenylcarbonyl)piperazin-2-yl]ethyl}-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Orth, P, Cumming, J. | | Deposit date: | 2010-02-02 | | Release date: | 2010-04-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Piperazine sulfonamide BACE1 inhibitors: design, synthesis, and in vivo characterization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
6D3K
 
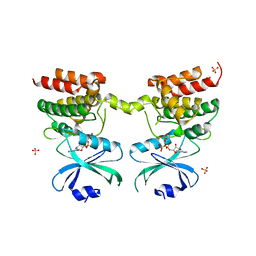 | | Crystal structure of unphosphorylated human PKR kinase domain in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Interferon-induced, double-stranded RNA-activated protein kinase, ... | | Authors: | Erlandsen, H, Mayo, C.B, Robinson, V.L, Cole, J.L. | | Deposit date: | 2018-04-16 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis of Protein Kinase R Autophosphorylation.
Biochemistry, 58, 2019
|
|
1IMX
 
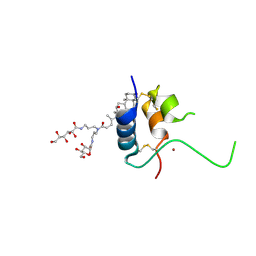 | | 1.8 Angstrom crystal structure of IGF-1 | | Descriptor: | BROMIDE ION, Insulin-like Growth Factor 1A, N,N-BIS(3-D-GLUCONAMIDOPROPYL)DEOXYCHOLAMIDE | | Authors: | Vajdos, F.F, Ultsch, M, Schaffer, M.L, Deshayes, K.D, Liu, J, Skelton, N.J, de Vos, A.M. | | Deposit date: | 2001-05-11 | | Release date: | 2001-09-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of human insulin-like growth factor-1: detergent binding inhibits binding protein interactions.
Biochemistry, 40, 2001
|
|
3L5D
 
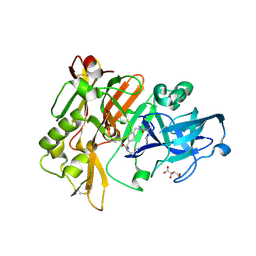 | | Structure of BACE Bound to SCH723873 | | Descriptor: | 1-butyl-3-(4-{[(2Z,4R)-2-imino-4-methyl-4-(2-methylpropyl)-5-oxoimidazolidin-1-yl]methyl}benzyl)urea, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
1ING
 
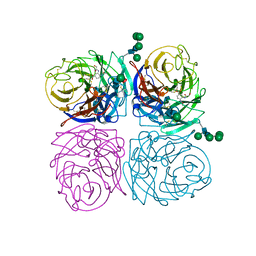 | | INFLUENZA A SUBTYPE N2 NEURAMINIDASE COMPLEXED WITH AROMATIC BANA109 INHIBITOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(ACETYLAMINO)-3-[(HYDROXYACETYL)AMINO]BENZOIC ACID, CALCIUM ION, ... | | Authors: | Jedrzejas, M.J, Luo, M. | | Deposit date: | 1995-07-07 | | Release date: | 1996-08-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based inhibitors of influenza virus sialidase. A benzoic acid lead with novel interaction.
J.Med.Chem., 38, 1995
|
|
3L9Y
 
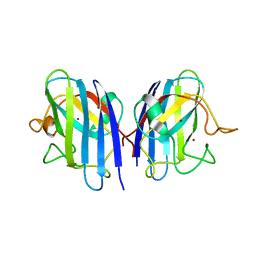 | | Crystal structures of holo and Cu-deficient Cu/ZnSOD from the silkworm Bombyx mori and the implications in Amyotrophic lateral sclerosis | | Descriptor: | COPPER (II) ION, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Zhang, N.-N, He, Y.-X, Li, W.-F, Zhao, F, Yan, L.-F, Zhang, G.-Z, Teng, Y.-B, Yu, J, Chen, Y, Zhou, C.-Z. | | Deposit date: | 2010-01-06 | | Release date: | 2010-03-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of holo and Cu-deficient Cu/Zn-SOD from the silkworm Bombyx mori and the implications in amyotrophic lateral sclerosis.
Proteins, 78, 2010
|
|
1IP2
 
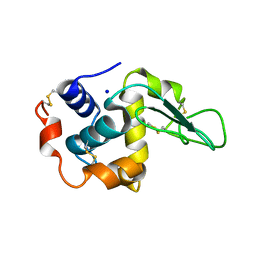 | | G48A HUMAN LYSOZYME | | Descriptor: | LYSOZYME C, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2001-04-20 | | Release date: | 2001-11-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of amino acid residues in left-handed helical conformation for the conformational stability of a protein.
Proteins, 45, 2001
|
|
6CWS
 
 | |
3LFA
 
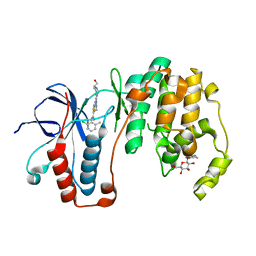 | | Human p38 MAP Kinase in Complex with Dasatinib | | Descriptor: | Mitogen-activated protein kinase 14, N-(2-CHLORO-6-METHYLPHENYL)-2-({6-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]-2-METHYLPYRIMIDIN-4-YL}AMINO)-1,3-THIAZOLE-5-CARBOXAMIDE, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Getlik, M, Rauh, D. | | Deposit date: | 2010-01-16 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Development of novel thiazole-urea compounds which stabalize the inactive conformation of p38 alpha
To be Published
|
|
3LAW
 
 | | Structure of GTP-bound L129F mutant Rab7 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein Rab-7a | | Authors: | McCray, B.A, Skordalakes, E, Taylor, J.P. | | Deposit date: | 2010-01-07 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Disease mutations in Rab7 result in unregulated nucleotide exchange and inappropriate activation.
Hum.Mol.Genet., 19, 2010
|
|
1IX0
 
 | | I59A-3SS human lysozyme | | Descriptor: | SODIUM ION, lysozyme | | Authors: | Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2002-06-06 | | Release date: | 2003-07-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Buried water molecules contribute to the conformational stability of a protein
PROTEIN ENG., 16, 2003
|
|
6CHA
 
 | |
1IZR
 
 | | F46A mutant of bovine pancreatic ribonuclease A | | Descriptor: | RIBONUCLEASE A | | Authors: | Kadonosono, T, Chatani, E, Hayashi, R, Moriyama, H, Ueki, T. | | Deposit date: | 2002-10-11 | | Release date: | 2003-11-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Minimization of cavity size ensures protein stability and folding: structures of Phe46-replaced bovine pancreatic RNase A
Biochemistry, 42, 2003
|
|
6D6H
 
 | | Triclinic lysozyme cryocooled to 100 K with 47% MPD as cryoprotectant | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Lysozyme C, NITRATE ION | | Authors: | Juers, D.H. | | Deposit date: | 2018-04-20 | | Release date: | 2018-09-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.000064 Å) | | Cite: | The impact of cryosolution thermal contraction on proteins and protein crystals: volumes, conformation and order.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
3LJJ
 
 | | Bovine trypsin in complex with UB-THR 10 | | Descriptor: | (S)-N-(4-carbamimidoylbenzyl)-1-(2-(cyclopentylamino)ethanoyl)pyrrolidine-2-carboxamide, CALCIUM ION, Cationic trypsin, ... | | Authors: | Wegscheid-Gerlach, C, Heine, A, Klebe, G. | | Deposit date: | 2010-01-26 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Congeneric but still distinct: how closely related trypsin ligands exhibit different thermodynamic and structural properties.
J.Mol.Biol., 405, 2011
|
|
