7ZLK
 
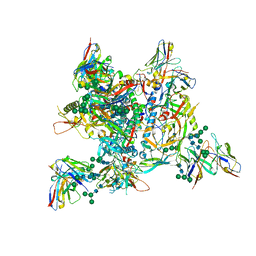 | | AMC009 SOSIPv5.2 in complex with Fabs ACS101 and ACS124 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACS114 heavy chain, ... | | Authors: | van Schooten, J, Ozorowski, G, Ward, A. | | Deposit date: | 2022-04-15 | | Release date: | 2022-09-21 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Complementary antibody lineages achieve neutralization breadth in an HIV-1 infected elite neutralizer.
Plos Pathog., 18, 2022
|
|
7ZJX
 
 | | Rabbit 80S ribosome programmed with SECIS and SBP2 | | Descriptor: | 18S rRNA, 28S rRNA, 40S Ribosomal protein eS19, ... | | Authors: | Hilal, T, Simonovic, M, Spahn, C.M.T. | | Deposit date: | 2022-04-12 | | Release date: | 2022-09-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the mammalian ribosome as it decodes the selenocysteine UGA codon.
Science, 376, 2022
|
|
7ZTS
 
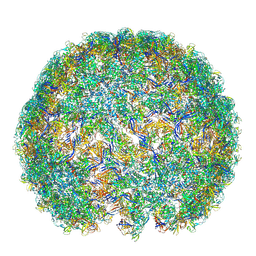 | | Saccharomyces cerevisiae L-BC virus, open particle, asymmetric reconstruction | | Descriptor: | Major capsid protein | | Authors: | Grybchuk, D, Prochazkova, M, Fuzik, T, Konovalovas, A, Serva, S, Yurchenko, V, Plevka, P. | | Deposit date: | 2022-05-11 | | Release date: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | Structures of L-BC virus and its open particle provide insight into Totivirus capsid assembly.
Commun Biol, 5, 2022
|
|
7ZXS
 
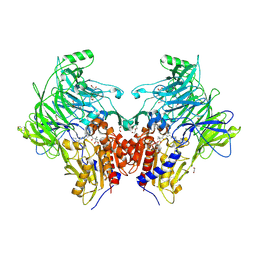 | | Crystal structure of DPP9 in complex with a 4-oxo-b-lactam based inhibitor, A295 | | Descriptor: | 1,2-ETHANEDIOL, 2-ethyl-2-methanoyl-~{N}-[3-[[4-(quinolin-8-ylmethyl)piperazin-1-yl]methyl]phenyl]butanamide, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ross, B, Huber, R. | | Deposit date: | 2022-05-22 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Chemoproteomics-Enabled Identification of 4-Oxo-beta-Lactams as Inhibitors of Dipeptidyl Peptidases 8 and 9.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
5JDW
 
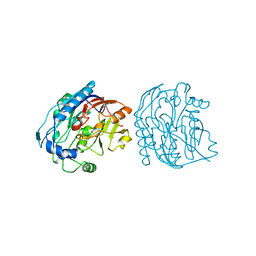 | |
5JGF
 
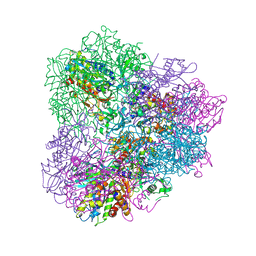 | | Crystal structure of mApe1 | | Descriptor: | Vacuolar aminopeptidase 1, ZINC ION | | Authors: | Noda, N.N, Adachi, W, Inagaki, F. | | Deposit date: | 2016-04-20 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Basis for Receptor-Mediated Selective Autophagy of Aminopeptidase I Aggregates
Cell Rep, 16, 2016
|
|
5YRS
 
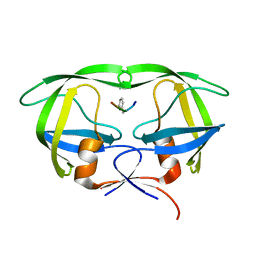 | | X-ray Snapshot of HIV-1 Protease in Action: Observation of Tetrahedral Intermediate and Its SIHB with Catalytic Aspartate | | Descriptor: | PROTEASE, RT-RH oligopeprtide | | Authors: | Das, A, Mahale, S, Prashar, V, Bihani, S, Ferrer, J.-L, Hosur, M.V. | | Deposit date: | 2017-11-10 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | X-ray snapshot of HIV-1 protease in action: observation of tetrahedral intermediate and short ionic hydrogen bond SIHB with catalytic aspartate.
J. Am. Chem. Soc., 132, 2010
|
|
4DCK
 
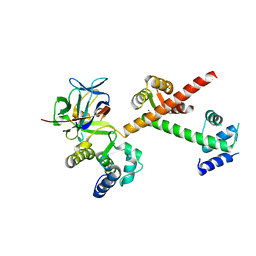 | | Crystal structure of the C-terminus of voltage-gated sodium channel in complex with FGF13 and CaM | | Descriptor: | Calmodulin, Fibroblast growth factor 13, MAGNESIUM ION, ... | | Authors: | Chung, B.C, Wang, C, Yan, H, Pitt, G.S, Lee, S.Y. | | Deposit date: | 2012-01-17 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Ternary Complex of a NaV C-Terminal Domain, a Fibroblast Growth Factor Homologous Factor, and Calmodulin.
Structure, 20, 2012
|
|
6PRC
 
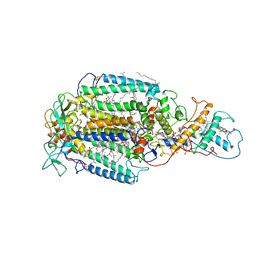 | | PHOTOSYNTHETIC REACTION CENTER FROM RHODOPSEUDOMONAS VIRIDIS (DG-420314 (TRIAZINE) COMPLEX) | | Descriptor: | 15-cis-1,2-dihydroneurosporene, 2-CHLORO-4-ETHYLAMINO-6-(S(-)-2'-CYANO-4-BUTYLAMINO)-1,3,5-TRIAZINE, BACTERIOCHLOROPHYLL B, ... | | Authors: | Lancaster, C.R.D, Michel, H. | | Deposit date: | 1997-07-31 | | Release date: | 1999-04-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Refined crystal structures of reaction centres from Rhodopseudomonas viridis in complexes with the herbicide atrazine and two chiral atrazine derivatives also lead to a new model of the bound carotenoid.
J.Mol.Biol., 286, 1999
|
|
5K0H
 
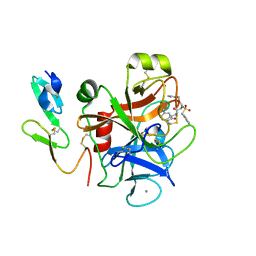 | |
8EST
 
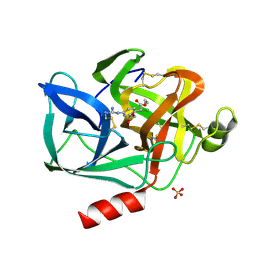 | | REACTION OF PORCINE PANCREATIC ELASTASE WITH 7-SUBSTITUTED 3-ALKOXY-4-CHLOROISOCOUMARINS: DESIGN OF POTENT INHIBITORS USING THE CRYSTAL STRUCTURE OF THE COMPLEX FORMED WITH 4-CHLORO-3-ETHOXY-7-GUANIDINO-ISOCOUMARIN | | Descriptor: | CALCIUM ION, ETHYL-(2-CARBOXY-4-GUANIDINIUM-PHENYL)-CHLOROACETATE, PORCINE PANCREATIC ELASTASE, ... | | Authors: | Radhakrishnan, R, Powers, J.C, Meyerjunior, E.F. | | Deposit date: | 1990-02-21 | | Release date: | 1992-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Reaction of porcine pancreatic elastase with 7-substituted 3-alkoxy-4-chloroisocoumarins: design of potent inhibitors using the crystal structure of the complex formed with 4-chloro-3-ethoxy-7-guanidinoisocoumarin.
Biochemistry, 29, 1990
|
|
4OR5
 
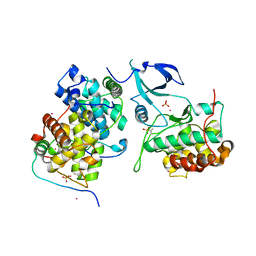 | | Crystal structure of HIV-1 Tat complexed with human P-TEFb and AFF4 | | Descriptor: | AF4/FMR2 family member 4, Cyclin-T1, Cyclin-dependent kinase 9, ... | | Authors: | Gu, J, Babayeva, N.D, Suwa, Y, Baranovskiy, A.G, Price, D.H, Tahirov, T.H. | | Deposit date: | 2014-02-10 | | Release date: | 2014-04-16 | | Last modified: | 2014-06-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of HIV-1 Tat complexed with human P-TEFb and AFF4.
Cell Cycle, 13, 2014
|
|
6Q21
 
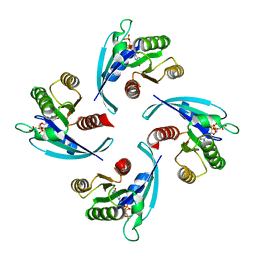 | |
5KNC
 
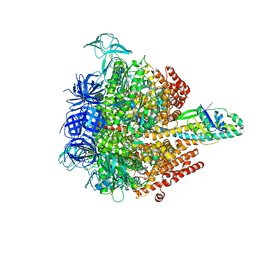 | | Crystal structure of the 3 ADP-bound V1 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Suzuki, K, Mizutani, K, Maruyama, S, Shimono, K, Imai, F.L, Muneyuki, E, Kakinuma, Y, Ishizuka-Katsura, Y, Shirouzu, M, Yokoyama, S, Yamato, I, Murata, T. | | Deposit date: | 2016-06-28 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.015 Å) | | Cite: | Crystal structures of the ATP-binding and ADP-release dwells of the V1 rotary motor
Nat Commun, 7, 2016
|
|
5J0A
 
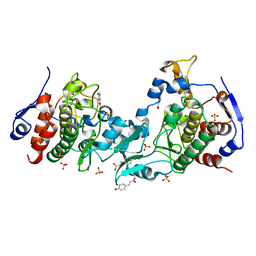 | | Crystal structure of PDZ-binding kinase | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Lymphokine-activated killer T-cell-originated protein kinase, SULFATE ION | | Authors: | Zou, Q.W, Zhou, H, Yang, X. | | Deposit date: | 2016-03-28 | | Release date: | 2016-09-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | The crystal structure of an inactive dimer of PDZ-binding kinase
Biochem.Biophys.Res.Commun., 476, 2016
|
|
4C8R
 
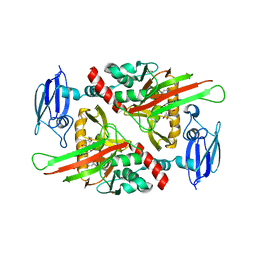 | | Human gamma-butyrobetaine dioxygenase (BBOX1) in complex with Ni(II) and N-(3-hydroxypicolinoyl)-S-(pyridin-2-ylmethyl)-L-cysteine (AR692B) | | Descriptor: | 1,2-ETHANEDIOL, GAMMA-BUTYROBETAINE DIOXYGENASE, N-(3-hydroxypicolinoyl)-S-(pyridin-2-ylmethyl)-L-cysteine, ... | | Authors: | Chowdhury, R, Rydzik, A.M, Kochan, G.T, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2013-10-01 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Modulating carnitine levels by targeting its biosynthesis pathway - selective inhibition of gamma-butyrobetaine hydroxylase.
Chem Sci, 5, 2014
|
|
5KVY
 
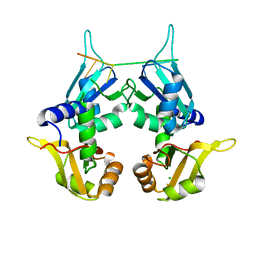 | | CRYSTAL STRUCTURE OF THE TWO TANDEM RRM DOMAINS OF PUF60 BOUND TO A PORTION OF AN ADML PRE-MRNA 3' SPLICE SITE ANALOG | | Descriptor: | CHLORIDE ION, DNA (30-MER), Poly(U)-binding-splicing factor PUF60 | | Authors: | Hsiao, H.-H, Crichlow, G.V, Albright, R.A, Murphy, J.W, Lolis, E.J, Braddock, D.T. | | Deposit date: | 2016-07-15 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Unraveling the mechanism of recognition of the 3' splice site of the adenovirus major late promoter intron by the alternative splicing factor PUF60.
Plos One, 15, 2020
|
|
6PTD
 
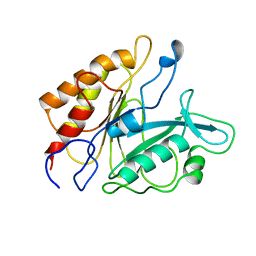 | | PHOSPHATIDYLINOSITOL-SPECIFIC PHOSPHOLIPASE C MUTANT H32L | | Descriptor: | PHOSPHATIDYLINOSITOL-SPECIFIC PHOSPHOLIPASE C | | Authors: | Heinz, D.W. | | Deposit date: | 1997-07-18 | | Release date: | 1998-01-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Probing the roles of active site residues in phosphatidylinositol-specific phospholipase C from Bacillus cereus by site-directed mutagenesis.
Biochemistry, 36, 1997
|
|
5KTQ
 
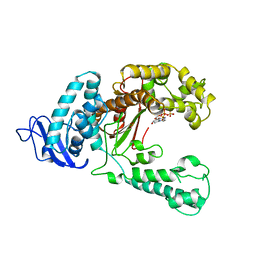 | | LARGE FRAGMENT OF TAQ DNA POLYMERASE BOUND TO DCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, PROTEIN (DNA POLYMERASE I) | | Authors: | Li, Y, Kong, Y, Korolev, S, Waksman, G. | | Deposit date: | 1998-09-22 | | Release date: | 1998-09-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the Klenow fragment of Thermus aquaticus DNA polymerase I complexed with deoxyribonucleoside triphosphates.
Protein Sci., 7, 1998
|
|
5Z5S
 
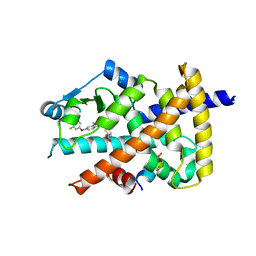 | | Crystal structure of the PPARgamma-LBD complexed with compound 13ab | | Descriptor: | 3-{[6-(4-chloro-3-fluorophenoxy)-1-methyl-1H-benzimidazol-2-yl]methoxy}benzoic acid, CHLORIDE ION, Peptide from Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, ... | | Authors: | Matsui, Y, Hanzawa, H. | | Deposit date: | 2018-01-19 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of DS-6930, a potent selective PPAR gamma modulator. Part I: Lead identification.
Bioorg. Med. Chem., 26, 2018
|
|
5WEE
 
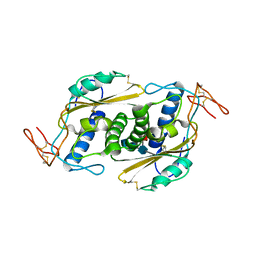 | | Crystal structure of HpVAL4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Asojo, O.A. | | Deposit date: | 2017-07-09 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Heligmosomoides polygyrus Venom Allergen-like Protein-4 (HpVAL-4) is a sterol binding protein.
Int. J. Parasitol., 48, 2018
|
|
5KNB
 
 | | Crystal structure of the 2 ADP-bound V1 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Suzuki, K, Mizutani, K, Maruyama, S, Shimono, K, Imai, F.L, Muneyuki, E, Kakinuma, Y, Ishizuka-Katsura, Y, Shirouzu, M, Yokoyama, S, Yamato, I, Murata, T. | | Deposit date: | 2016-06-28 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.251 Å) | | Cite: | Crystal structures of the ATP-binding and ADP-release dwells of the V1 rotary motor
Nat Commun, 7, 2016
|
|
5KND
 
 | | Crystal structure of the Pi-bound V1 complex | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Suzuki, K, Mizutani, K, Maruyama, S, Shimono, K, Imai, F.L, Muneyuki, E, Kakinuma, Y, Ishizuka-Katsura, Y, Shirouzu, M, Yokoyama, S, Yamato, I, Murata, T. | | Deposit date: | 2016-06-28 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.888 Å) | | Cite: | Crystal structures of the ATP-binding and ADP-release dwells of the V1 rotary motor
Nat Commun, 7, 2016
|
|
5Z6S
 
 | | Crystal structure of the PPARgamma-LBD complexed with compound DS-6930 | | Descriptor: | 3-[[6-(3,5-dimethylpyridin-2-yl)oxy-1-methyl-benzimidazol-2-yl]methoxy]benzoic acid, CHLORIDE ION, Peptide from Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, ... | | Authors: | Matsui, Y, Hanzawa, H. | | Deposit date: | 2018-01-25 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of DS-6930, a potent selective PPAR gamma modulator. Part II: Lead optimization.
Bioorg. Med. Chem., 26, 2018
|
|
5KWQ
 
 | | Two Tandem RRM Domains of FBP-Interacting Repressor (FIR), also Known as PUF60 | | Descriptor: | Poly(U)-binding-splicing factor PUF60 | | Authors: | Crichlow, G.V, Yang, Y, Zhou, H, Lolis, E.J, Braddock, D.T. | | Deposit date: | 2016-07-18 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Unraveling the mechanism of recognition of the 3' splice site of the adenovirus major late promoter intron by the alternative splicing factor PUF60.
Plos One, 15, 2020
|
|
