1GU5
 
 | |
1GTW
 
 | |
7RKP
 
 | | The crystal structure of I38T mutant PA endonuclease (2009/H1N1/CALIFORNIA) in complex with cyclic compound SJ001034733 | | Descriptor: | (5R)-5-hydroxy-16,21-dioxa-3,8,28-triazatetracyclo[20.3.1.1~2,6~.1~11,15~]octacosa-1(26),2,6(28),11(27),12,14,22,24-octaene-4,7-dione, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, J.P, Rankovic, Z, White, S.W. | | Deposit date: | 2021-07-22 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
5T7B
 
 | |
6K6S
 
 | |
2XB2
 
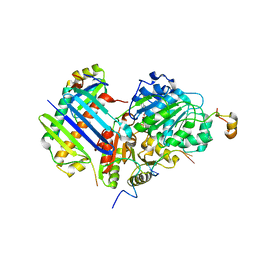 | | Crystal structure of the core Mago-Y14-eIF4AIII-Barentsz-UPF3b assembly shows how the EJC is bridged to the NMD machinery | | Descriptor: | EUKARYOTIC INITIATION FACTOR 4A-III, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Buchwald, G, Ebert, J, Basquin, C, Sauliere, J, Jayachandran, U, Bono, F, Le Hir, H, Conti, E. | | Deposit date: | 2010-04-03 | | Release date: | 2010-05-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Insights Into the Recruitment of the Nmd Machinery from the Crystal Structure of a Core Ejc-Upf3B Complex.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2ZJP
 
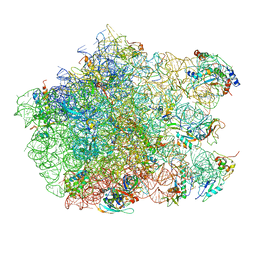 | | Thiopeptide antibiotic Nosiheptide bound to the large ribosomal subunit of Deinococcus radiodurans | | Descriptor: | 4-(hydroxymethyl)-3-methyl-1H-indole-2-carboxylic acid, 50S RIBOSOMAL PROTEIN L11, 50S RIBOSOMAL PROTEIN L13, ... | | Authors: | Harms, J.M, Wilson, D.N, Schluenzen, F, Connell, S.R, Stachelhaus, T, Zaborowska, Z, Spahn, C.M.T, Fucini, P. | | Deposit date: | 2008-03-07 | | Release date: | 2008-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Translational Regulation Via L11: Molecular Switches on the Ribosome Turned on and Off by Thiostrepton and Micrococcin.
Mol.Cell, 30, 2008
|
|
7X76
 
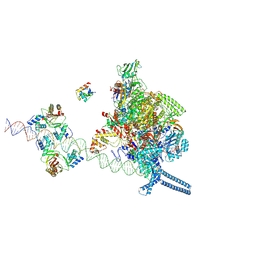 | |
7X75
 
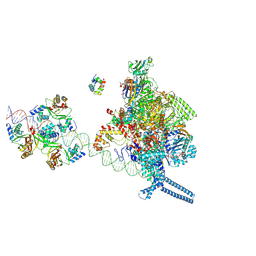 | |
1R0V
 
 | |
1R11
 
 | |
3VF0
 
 | | Raver1 in complex with metavinculin L954 deletion mutant | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Ribonucleoprotein PTB-binding 1, ... | | Authors: | Lee, J.H, Vonrhein, C, Bricogne, G, Izard, T. | | Deposit date: | 2012-01-09 | | Release date: | 2012-07-25 | | Last modified: | 2012-09-19 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | The Metavinculin Tail Domain Directs Constitutive Interactions with Raver1 and vinculin RNA.
J.Mol.Biol., 422, 2012
|
|
5MKO
 
 | | [2Fe-2S] cluster containing TtuA in complex with AMP. | | Descriptor: | ADENOSINE MONOPHOSPHATE, FE2/S2 (INORGANIC) CLUSTER, TtuA PH0300, ... | | Authors: | Arragain, S, Bimai, O, Legrand, P, Golinelli-Pimpaneau, B. | | Deposit date: | 2016-12-05 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Nonredox thiolation in tRNA occurring via sulfur activation by a [4Fe-4S] cluster.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7W89
 
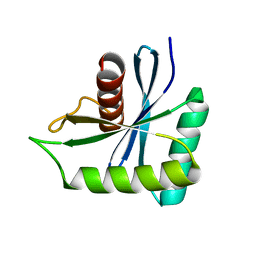 | | The structure of Deinococcus radiodurans Yqgf | | Descriptor: | Putative pre-16S rRNA nuclease | | Authors: | Cheng, K. | | Deposit date: | 2021-12-07 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.500061 Å) | | Cite: | Biochemical and Structural Study of RuvC and YqgF from Deinococcus radiodurans.
Mbio, 13, 2022
|
|
2C77
 
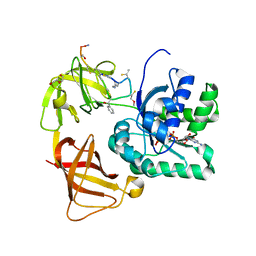 | | EF-Tu complexed with a GTP analog and the antibiotic GE2270 A | | Descriptor: | DI(HYDROXYETHYL)ETHER, ELONGATION FACTOR TU-B, MAGNESIUM ION, ... | | Authors: | Parmeggiani, A, Krab, I.M, Okamura, S, Nielsen, R.C, Nyborg, J, Nissen, P. | | Deposit date: | 2005-11-18 | | Release date: | 2006-03-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of the action of pulvomycin and GE2270 A on elongation factor Tu.
Biochemistry, 45, 2006
|
|
7P5X
 
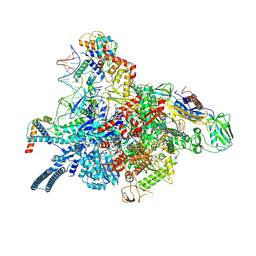 | | Mycobacterial RNAP with transcriptional activator PafBC | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Mueller, A.U, Kummer, E, Schilling, C.M, Ban, N, Weber-Ban, E. | | Deposit date: | 2021-07-15 | | Release date: | 2021-12-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Transcriptional control of mycobacterial DNA damage response by sigma adaptation.
Sci Adv, 7, 2021
|
|
5DIL
 
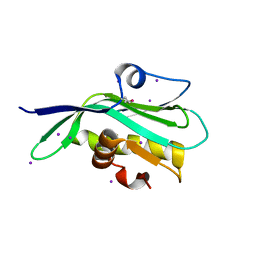 | | Crystal structure of the effector domain of the NS1 protein from influenza virus B | | Descriptor: | IODIDE ION, Non-structural protein 1 | | Authors: | Guan, R, Hamilton, K, Ma, L, Montelione, G.T. | | Deposit date: | 2015-09-01 | | Release date: | 2016-08-10 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A Second RNA-Binding Site in the NS1 Protein of Influenza B Virus.
Structure, 24, 2016
|
|
5LVF
 
 | | Solution structure of Rtt103 CTD-interacting domain bound to a Thr4 phosphorylated CTD peptide | | Descriptor: | PRO-SER-TYR-SER-PRO-PTH-SER-PRO-SER-TYR-SER-PRO-THR-SER-PRO-SER, Regulator of Ty1 transposition protein 103 | | Authors: | Jasnovidova, O, Kubicek, K, Krejcikova, M, Stefl, R. | | Deposit date: | 2016-09-14 | | Release date: | 2017-05-10 | | Last modified: | 2019-10-30 | | Method: | SOLUTION NMR | | Cite: | Structural insight into recognition of phosphorylated threonine-4 of RNA polymerase II C-terminal domain by Rtt103p.
EMBO Rep., 18, 2017
|
|
5DY9
 
 | | Y68T Hfq from Methanococcus jannaschii in complex with AMP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Nikulin, A.D, Mikhailina, A.O, Lekontseva, N.V, Balobanov, V.A, Nikonova, E.Y, Tishchenko, S.V. | | Deposit date: | 2015-09-24 | | Release date: | 2016-09-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Characterization of RNA-binding properties of the archaeal Hfq-like protein from Methanococcus jannaschii.
J. Biomol. Struct. Dyn., 35, 2017
|
|
3CC4
 
 | |
2EHG
 
 | |
8GBD
 
 | | Homo sapiens Zalpha mutant - P193A | | Descriptor: | Double-stranded RNA-specific adenosine deaminase | | Authors: | Langeberg, C.J, Vogeli, B, Nichols, P.J, Henen, M, Vicens, Q. | | Deposit date: | 2023-02-25 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Differential Structural Features of Two Mutant ADAR1p150 Z alpha Domains Associated with Aicardi-Goutieres Syndrome.
J.Mol.Biol., 435, 2023
|
|
8GBC
 
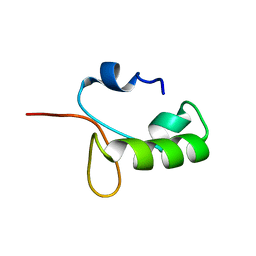 | | Homo sapiens Zalpha mutant - N173S | | Descriptor: | Double-stranded RNA-specific adenosine deaminase | | Authors: | Langeberg, C.J, Nichols, P.J, Henen, M, Vicens, Q, Vogeli, B. | | Deposit date: | 2023-02-25 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Differential Structural Features of Two Mutant ADAR1p150 Z alpha Domains Associated with Aicardi-Goutieres Syndrome.
J.Mol.Biol., 435, 2023
|
|
8RC4
 
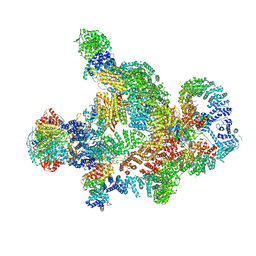 | | Structure of Integrator-PP2A complex | | Descriptor: | DSS1, Integrator complex subunit 1, Integrator complex subunit 10, ... | | Authors: | Fianu, I, Ochmann, M, Walshe, J.L, Cramer, P. | | Deposit date: | 2023-12-06 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of Integrator-dependent RNA polymerase II termination.
Nature, 629, 2024
|
|
8G9N
 
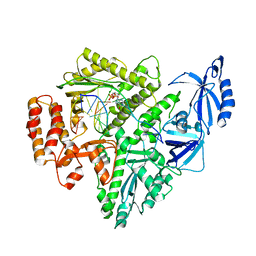 | | Partial DNA elongation subcomplex of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA template, ... | | Authors: | Mullins, E.A, Durie, C.L, Ohi, M.D, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
