5HBB
 
 | | Crystal structure of shaft pilin spaA from Lactobacillus rhamnosus GG - E139A mutant | | Descriptor: | 1,2-ETHANEDIOL, Cell surface protein SpaA, SODIUM ION, ... | | Authors: | Chaurasia, P, Pratap, S, von Ossowski, I, Palva, A, Krishnan, V. | | Deposit date: | 2015-12-31 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | New insights about pilus formation in gut-adapted Lactobacillus rhamnosus GG from the crystal structure of the SpaA backbone-pilin subunit
Sci Rep, 6, 2016
|
|
5Z72
 
 | |
4TVO
 
 | |
6TBF
 
 | | Structure of a beta galactosidase with inhibitor | | Descriptor: | (1~{S},2~{S},3~{S},4~{R},5~{R})-4-(hydroxymethyl)-5-(octylamino)cyclopentane-1,2,3-triol, ACETATE ION, Beta-galactosidase, ... | | Authors: | Offen, W, Davies, G. | | Deposit date: | 2019-11-01 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanistic Insights into the Chaperoning of Human Lysosomal-Galactosidase Activity: Highly Functionalized Aminocyclopentanes and C -5a-Substituted Derivatives of 4- epi -Isofagomine.
Molecules, 25, 2020
|
|
2OTL
 
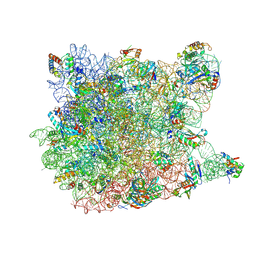 | | Girodazole bound to the large subunit of Haloarcula marismortui | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L10E, 50S ribosomal protein L10e, ... | | Authors: | Blaha, G, Schroeder, S.J, Tirado-Rives, J. | | Deposit date: | 2007-02-08 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Structures of Antibiotics Bound to the E Site Region of the 50 S Ribosomal Subunit of Haloarcula marismortui: 13-Deoxytedanolide and Girodazole.
J.Mol.Biol., 367, 2007
|
|
5G3P
 
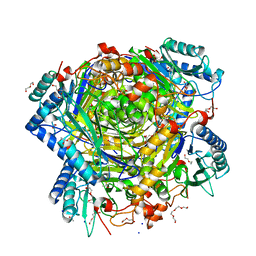 | | Bacillus cereus formamidase (BceAmiF) acetylated at the active site. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gavira, J.A, Conejero-Muriel, M, Martinez-Rodriguez, S. | | Deposit date: | 2016-04-29 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A novel cysteine carbamoyl-switch is responsible for the inhibition of formamidase, a nitrilase superfamily member.
Arch.Biochem.Biophys., 662, 2019
|
|
2OTJ
 
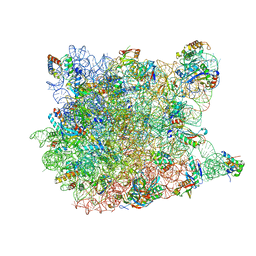 | | 13-deoxytedanolide bound to the large subunit of Haloarcula marismortui | | Descriptor: | 13-DEOXYTEDANOLIDE, 23S ribosomal RNA, 50S ribosomal protein L10E, ... | | Authors: | Blaha, G, Schroeder, S.J, Tirado-Rives, J. | | Deposit date: | 2007-02-08 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Structures of Antibiotics Bound to the E Site Region of the 50 S Ribosomal Subunit of Haloarcula marismortui: 13-Deoxytedanolide and Girodazole.
J.Mol.Biol., 367, 2007
|
|
6KD1
 
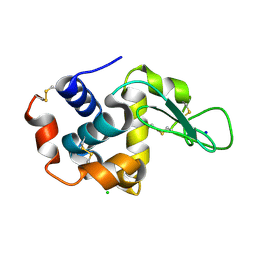 | |
6KOK
 
 | | Crystal Structure of SNX11/SNX10-PXe Chimera | | Descriptor: | CHLORIDE ION, SODIUM ION, Sorting nexin-11,Uncharacterized protein SNX10 | | Authors: | Xu, T, Liu, J. | | Deposit date: | 2019-08-11 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Basis for PI(3,5)P2Recognition by SNX11, a Protein Involved in Lysosomal Degradation and Endosome Homeostasis Regulation.
J.Mol.Biol., 432, 2020
|
|
6KTW
 
 | | structure of EanB with hercynine | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wu, L, Liu, P.H, Zhou, J.H. | | Deposit date: | 2019-08-29 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.931 Å) | | Cite: | Single-Step Replacement of an Unreactive C-H Bond by a C-S Bond Using Polysulfide as the Direct Sulfur Source in the Anaerobic Ergothioneine Biosynthesis
Acs Catalysis, 10, 2020
|
|
6KQU
 
 | | Crystal structure of phospholipase A2 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hou, S, Xu, T, Liu, J. | | Deposit date: | 2019-08-18 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Residue Asn21 acts as a switch for calcium binding to modulate the enzymatic activity of human phospholipase A2 group IIE.
Biochimie, 176, 2020
|
|
6L9C
 
 | | Neutron structure of copper amine oxidase from Arthrobacter glibiformis at pD 7.4 | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase, SODIUM ION | | Authors: | Murakawa, T, Kurihara, K, Shoji, M, Shibazaki, C, Sunami, T, Tamada, T, Yano, N, Yamada, T, Kusaka, K, Suzuki, M, Shigeta, Y, Kuroki, R, Hayashi, H, Yano, Y, Tanizawa, K, Adachi, M, Okajima, T. | | Deposit date: | 2019-11-08 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-22 | | Method: | NEUTRON DIFFRACTION (1.14 Å), X-RAY DIFFRACTION | | Cite: | Neutron crystallography of copper amine oxidase reveals keto/enolate interconversion of the quinone cofactor and unusual proton sharing.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6L18
 
 | |
6L4P
 
 | | Crystal structure of the complex between the axonemal outer-arm dynein light chain-1 and microtubule binding domain of gamma heavy chain | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Dynein light chain 1, axonemal, ... | | Authors: | Toda, A, Nishikawa, Y, Tanaka, H, Yagi, T, Kurisu, G. | | Deposit date: | 2019-10-19 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | The complex of outer-arm dynein light chain-1 and the microtubule-binding domain of the gamma heavy chain shows how axonemal dynein tunes ciliary beating.
J.Biol.Chem., 295, 2020
|
|
6KV0
 
 | | Ferredoxin I from C. reinhardtii, high X-ray dose | | Descriptor: | BENZAMIDINE, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Onishi, Y, Kurisu, G, Tanaka, H. | | Deposit date: | 2019-09-03 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray dose-dependent structural changes of the [2Fe-2S] ferredoxin from Chlamydomonas reinhardtii.
J.Biochem., 167, 2020
|
|
6KUM
 
 | | Ferredoxin I from C. reinhardtii, low X-ray dose | | Descriptor: | BENZAMIDINE, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Onishi, Y, Kurisu, G, Tanaka, H. | | Deposit date: | 2019-09-02 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray dose-dependent structural changes of the [2Fe-2S] ferredoxin from Chlamydomonas reinhardtii.
J.Biochem., 167, 2020
|
|
6LBL
 
 | | Crystal structure of IMP-1 metallo-beta-lactamase in complex with NO9 inhibitor | | Descriptor: | 2,5-dimethyl-4-sulfamoyl-furan-3-carboxylic acid, Metallo-beta-lactamase type 2, SODIUM ION, ... | | Authors: | Wachino, J. | | Deposit date: | 2019-11-14 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Sulfamoyl Heteroarylcarboxylic Acids as Promising Metallo-beta-Lactamase Inhibitors for Controlling Bacterial Carbapenem Resistance.
Mbio, 11, 2020
|
|
6LFW
 
 | |
8E6K
 
 | | 2H08 Fab in complex with influenza virus neuraminidase from A/Brevig Mission/1/1918 (H1N1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2H08 fragment antigen binding heavy chain, 2H08 fragment antigen binding light chain, ... | | Authors: | Turner, H.L, Ozorowski, G, Ward, A.B. | | Deposit date: | 2022-08-22 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Human anti-N1 monoclonal antibodies elicited by pandemic H1N1 virus infection broadly inhibit HxN1 viruses in vitro and in vivo.
Immunity, 56, 2023
|
|
8E6J
 
 | |
5GS8
 
 | | Crystal structure of TLA-3 extended-spectrum beta-lactamase | | Descriptor: | Beta-lactamase, CHLORIDE ION, SODIUM ION, ... | | Authors: | Wachino, J, Jin, W, Arakawa, Y. | | Deposit date: | 2016-08-14 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural Insights into the TLA-3 Extended-Spectrum beta-Lactamase and Its Inhibition by Avibactam and OP0595.
Antimicrob. Agents Chemother., 61, 2017
|
|
5C3Y
 
 | | Structure of human ribokinase crystallized with AMPPNP | | Descriptor: | AMP PHOSPHORAMIDATE, Ribokinase, SODIUM ION | | Authors: | Park, J, Chakrabarti, J, Singh, B, Gupta, R.S, Junop, M.S. | | Deposit date: | 2015-06-17 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of human ribokinase crystallized with AMPPNP
To Be Published
|
|
6B5S
 
 | | Structure of PfCSP peptide 25 with human antibody CIS42 | | Descriptor: | AMMONIUM ION, CIS42 Fab Heavy chain, CIS42 Fab Light chain, ... | | Authors: | Pancera, M, Weidle, C. | | Deposit date: | 2017-09-29 | | Release date: | 2018-03-21 | | Last modified: | 2019-01-09 | | Method: | X-RAY DIFFRACTION (1.983 Å) | | Cite: | A human monoclonal antibody prevents malaria infection by targeting a new site of vulnerability on the parasite.
Nat. Med., 24, 2018
|
|
6TBJ
 
 | | Structure of a beta galactosidase with inhibitor | | Descriptor: | 5-(dimethylamino)-~{N}-[6-[(2~{S},3~{R},4~{S},5~{R})-3-(hydroxymethyl)-4,5-bis(oxidanyl)piperidin-2-yl]hexyl]naphthalene-1-sulfonamide, Beta-galactosidase, putative, ... | | Authors: | Offen, W, Davies, G. | | Deposit date: | 2019-11-01 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanistic Insights into the Chaperoning of Human Lysosomal-Galactosidase Activity: Highly Functionalized Aminocyclopentanes and C -5a-Substituted Derivatives of 4- epi -Isofagomine.
Molecules, 25, 2020
|
|
6TBH
 
 | | Structure of a beta galactosidase with inhibitor | | Descriptor: | 5-[ethyl(methyl)amino]-~{N}-[6-[[(1~{S},2~{R},3~{S},4~{R})-2-(hydroxymethyl)-3,4-bis(oxidanyl)cyclopentyl]amino]hexyl]naphthalene-1-sulfonamide, ACETATE ION, Beta-galactosidase, ... | | Authors: | Offen, W, Davies, G. | | Deposit date: | 2019-11-01 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanistic Insights into the Chaperoning of Human Lysosomal-Galactosidase Activity: Highly Functionalized Aminocyclopentanes and C -5a-Substituted Derivatives of 4- epi -Isofagomine.
Molecules, 25, 2020
|
|
