4W4G
 
 | | Postcleavage state of 70S bound to HigB toxin and AAA (lysine) codon | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Schureck, M.A, Maehigashi, T, Dunkle, J.A, Dunham, C.M. | | Deposit date: | 2014-08-14 | | Release date: | 2015-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Defining the mRNA recognition signature of a bacterial toxin protein.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4V95
 
 | | Crystal structure of YAEJ bound to the 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Gagnon, M.G, Seetharaman, S.V, Bulkley, D.P, Steitz, T.A. | | Deposit date: | 2012-01-27 | | Release date: | 2014-07-09 | | Last modified: | 2018-07-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the rescue of stalled ribosomes: structure of YaeJ bound to the ribosome.
Science, 335, 2012
|
|
4V41
 
 | | E. COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) | | Descriptor: | BETA-GALACTOSIDASE, MAGNESIUM ION | | Authors: | Juers, D.H, Jacobson, R.H, Wigley, D, Zhang, X.J, Huber, R.E, Tronrud, D.E, Matthews, B.W. | | Deposit date: | 2000-06-07 | | Release date: | 2014-07-09 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation.
Protein Sci., 9, 2000
|
|
4V4R
 
 | | Crystal structure of the whole ribosomal complex. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Petry, S, Brodersen, D.E, Murphy IV, F.V, Dunham, C.M, Selmer, M, Tarry, M.J, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2005-09-30 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (5.9 Å) | | Cite: | Crystal Structures of the Ribosome in Complex with Release Factors RF1 and RF2 Bound to a Cognate Stop Codon.
Cell(Cambridge,Mass.), 123, 2005
|
|
4V53
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli in complex with gentamicin. | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, 16S rRNA, 23S rRNA, ... | | Authors: | Borovinskaya, M.A, Pai, R.D, Zhang, W, Schuwirth, B.-S, Holton, J.M, Hirokawa, G, Kaji, H, Kaji, A, Cate, J.H.D. | | Deposit date: | 2007-06-16 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.54 Å) | | Cite: | Structural basis for aminoglycoside inhibition of bacterial ribosome recycling.
Nat.Struct.Mol.Biol., 14, 2007
|
|
4V6D
 
 | | Crystal structure of the E. coli 70S ribosome in an intermediate state of ratcheting | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, W, Dunkle, J.A, Cate, J.H.D. | | Deposit date: | 2009-06-27 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3.814 Å) | | Cite: | Structures of the ribosome in intermediate States of ratcheting.
Science, 325, 2009
|
|
4V6Y
 
 | | E. coli 70S-fMetVal-tRNAVal-tRNAfMet complex in classic pre-translocation state (pre1a) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Blau, C, Bock, L.V, Schroder, G.F, Davydov, I, Fischer, N, Stark, H, Rodnina, M.V, Vaiana, A.C, Grubmuller, H. | | Deposit date: | 2013-10-14 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Energy barriers and driving forces in tRNA translocation through the ribosome.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4V7C
 
 | | Structure of the Ribosome with Elongation Factor G Trapped in the Pre-Translocation State (pre-translocation 70S*tRNA structure) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Brilot, A.F, Korostelev, A.A, Ermolenko, D.N, Grigorieff, N. | | Deposit date: | 2013-11-20 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structure of the ribosome with elongation factor G trapped in the pretranslocation state.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3LE6
 
 | | The structure of cyclin dependent kinase 2 (CKD2) with a pyrazolobenzodiazepine inhibitor | | Descriptor: | 5-(2-chlorophenyl)-3-methyl-7-nitropyrazolo[3,4-b][1,4]benzodiazepine, Cell division protein kinase 2 | | Authors: | Lukacs, C.M, Swain, A, Crowther, R.L, Kammlott, R.U, Liu, J.J. | | Deposit date: | 2010-01-14 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pyrazolobenzodiazepines: part I. Synthesis and SAR of a potent class of kinase inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3LYX
 
 | | Crystal structure of the PAS domain of the protein CPS_1291 from Colwellia psychrerythraea. Northeast Structural Genomics Consortium target id CsR222B | | Descriptor: | Sensory box/GGDEF domain protein | | Authors: | Seetharaman, J, Chen, Y, Wang, D, Janjua, H, Cunningham, K, Owens, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-02-28 | | Release date: | 2010-04-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the PAS domain of the protein CPS_1291 from Colwellia psychrerythraea. Northeast Structural Genomics Consortium target id CsR222B
To be Published
|
|
3LW3
 
 | |
3LYO
 
 | | CROSS-LINKED CHICKEN LYSOZYME CRYSTAL IN 95% ACETONITRILE-WATER | | Descriptor: | ACETONITRILE, LYSOZYME | | Authors: | Huang, Q, Wang, Z, Zhu, G, Qian, M, Shao, M, Jia, Y, Tang, Y. | | Deposit date: | 1998-03-11 | | Release date: | 1998-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | X-ray studies on cross-linked lysozyme crystals in acetonitrile-water mixture.
Biochim.Biophys.Acta, 1384, 1998
|
|
3LZ2
 
 | | STRUCTURE DETERMINATION OF TURKEY EGG WHITE LYSOZYME USING LAUE DIFFRACTION | | Descriptor: | TURKEY EGG WHITE LYSOZYME | | Authors: | Howell, P.L, Almo, S.C, Parsons, M.R, Hajdu, J, Petsko, G.A. | | Deposit date: | 1991-09-13 | | Release date: | 1993-10-31 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure determination of turkey egg-white lysozyme using Laue diffraction data.
Acta Crystallogr.,Sect.B, 48, 1992
|
|
3LYS
 
 | | Crystal Structure of the N-terminal domain of the Prophage pi2 protein 01 (integrase) from Lactococcus lactis, Northeast Structural Genomics Consortium Target KR124F | | Descriptor: | Prophage pi2 protein 01, integrase | | Authors: | Forouhar, F, Abashidze, M, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Belote, R.L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-02-28 | | Release date: | 2010-03-16 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Northeast Structural Genomics Consortium Target KR124F
To be Published
|
|
3LAD
 
 | |
3MIN
 
 | | NITROGENASE MOFE PROTEIN FROM AZOTOBACTER VINELANDII, OXIDIZED STATE | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Peters, J.W, Stowell, M.H.B, Soltis, S.M, Day, M.W, Kim, J, Rees, D.C. | | Deposit date: | 1996-12-20 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Redox-dependent structural changes in the nitrogenase P-cluster.
Biochemistry, 36, 1997
|
|
3MAG
 
 | | VACCINIA METHYLTRANSFERASE VP39 COMPLEXED WITH M3ADE AND S-ADENOSYLHOMOCYSTEINE | | Descriptor: | 6-AMINO-3-METHYLPURINE, S-ADENOSYL-L-HOMOCYSTEINE, VP39 | | Authors: | Hu, G, Hodel, A.E, Gershon, P.D, Quiocho, F.A. | | Deposit date: | 1998-07-12 | | Release date: | 1999-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | mRNA cap recognition: dominant role of enhanced stacking interactions between methylated bases and protein aromatic side chains.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
3MCT
 
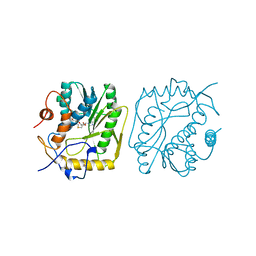 | | VACCINIA METHYLTRANSFERASE VP39 COMPLEXED WITH M3CYT AND S-ADENOSYLHOMOCYSTEINE | | Descriptor: | 3-METHYLCYTOSINE, S-ADENOSYL-L-HOMOCYSTEINE, VP39 | | Authors: | Hu, G, Gershon, P.D, Hodel, A.E, Quiocho, F.A. | | Deposit date: | 1999-01-05 | | Release date: | 1999-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | mRNA cap recognition: dominant role of enhanced stacking interactions between methylated bases and protein aromatic side chains.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
3M0X
 
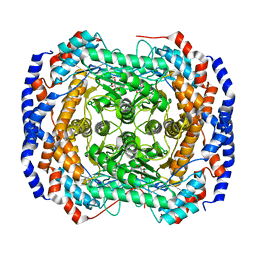 | | Crystal structure of Pseudomonas stutzeri L-rhamnose isomerase mutant S329L in complex with D-psicose | | Descriptor: | D-psicose, L-rhamnose isomerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Takeda, K, Izumori, K, Kamitori, S. | | Deposit date: | 2010-03-03 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Elucidation of the role of Ser329 and the C-terminal region in the catalytic activity of Pseudomonas stutzeri L-rhamnose isomerase
Protein Eng.Des.Sel., 23, 2010
|
|
4UX2
 
 | | Cryo-EM structure of antagonist-bound E2P gastric H,K-ATPase (SCH.E2. MgF) | | Descriptor: | POTASSIUM-TRANSPORTING ATPASE ALPHA CHAIN 1, POTASSIUM-TRANSPORTING ATPASE SUBUNIT BETA | | Authors: | Abe, K, Tani, K, Fujiyoshi, Y. | | Deposit date: | 2014-08-18 | | Release date: | 2014-09-17 | | Last modified: | 2014-11-12 | | Method: | ELECTRON CRYSTALLOGRAPHY (7 Å) | | Cite: | Systematic Comparison of Molecular Conformations of H+,K+-ATPase Reveals an Important Contribution of the A-M2 Linker for the Luminal Gating.
J.Biol.Chem., 289, 2014
|
|
4V5Q
 
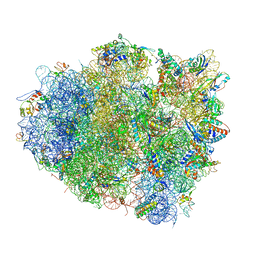 | | The crystal structure of EF-Tu and G24A-tRNA-Trp bound to a near- cognate codon on the 70S ribosome | | Descriptor: | 16S RRNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Schmeing, T.M, Voorhees, R.M, Ramakrishnan, V. | | Deposit date: | 2010-12-07 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | How Mutations in tRNA Distant from the Anticodon Affect the Fidelity of Decoding.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3MO0
 
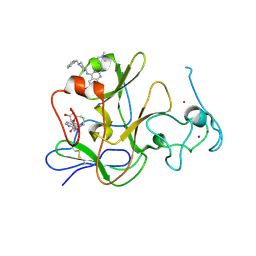 | | Human G9a-like (GLP, also known as EHMT1) in complex with inhibitor E11 | | Descriptor: | 1,2-ETHANEDIOL, Histone-lysine N-methyltransferase, H3 lysine-9 specific 5, ... | | Authors: | Chang, Y, Horton, J.R, Cheng, X. | | Deposit date: | 2010-04-22 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Adding a lysine mimic in the design of potent inhibitors of histone lysine methyltransferases.
J.Mol.Biol., 400, 2010
|
|
3MP1
 
 | | Complex structure of Sgf29 and trimethylated H3K4 | | Descriptor: | ACETATE ION, H3K4me3 peptide, Maltose-binding periplasmic protein,LINKER,SAGA-associated factor 29, ... | | Authors: | Li, J, Ruan, J, Wu, M, Xue, X, Zang, J. | | Deposit date: | 2010-04-24 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation
Embo J., 30, 2011
|
|
3MOW
 
 | | Crystal structure of SHP2 in complex with a tautomycetin analog TTN D-1 | | Descriptor: | (2Z)-2-[(1R)-3-{[(1R,2S,3R,6S,7S,10S,12S,15E,17E)-18-carboxy-16-ethyl-3,7-dihydroxy-1,2,6,10,12-pentamethyl-5-oxooctade ca-15,17-dien-1-yl]oxy}-1-hydroxy-3-oxopropyl]-3-methylbut-2-enedioic acid, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Zhang, Z.-Y, Liu, S, Yu, Z, Yu, X. | | Deposit date: | 2010-04-23 | | Release date: | 2011-05-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of SHP2 in complex with a tautomycetin analog TTN D-1
To be Published
|
|
4V90
 
 | | Thermus thermophilus Ribosome | | Descriptor: | 16S RIBOSOMAL RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Chen, Y, Feng, S, Kumar, V, Ero, R, Gao, Y.G. | | Deposit date: | 2014-02-22 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of EF-G-ribosome complex in a pretranslocation state.
Nat. Struct. Mol. Biol., 20, 2013
|
|
