5KGH
 
 | | X-ray structure of a glucosamine N-Acetyltransferase from Clostridium acetobutylicum, mutant Y297F | | Descriptor: | 1,2-ETHANEDIOL, ACETYL COENZYME *A, CHLORIDE ION, ... | | Authors: | Dopkins, B.J, Thoden, J.B, Tipton, P.A, Holden, H.M. | | Deposit date: | 2016-06-13 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Studies on a Glucosamine/Glucosaminide N-Acetyltransferase.
Biochemistry, 55, 2016
|
|
3KG0
 
 | | Crystal structure of SnoaB, a cofactor-independent oxygenase from Streptomyces nogalater, determined to 1.7 resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SnoaB | | Authors: | Koskiniemi, H, Grocholski, T, Lindqvist, Y, Mantsala, P, Niemi, J, Schneider, G. | | Deposit date: | 2009-10-28 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the cofactor-independent monooxygenase SnoaB from Streptomyces nogalater: implications for the reaction mechanism
Biochemistry, 49, 2010
|
|
3SNZ
 
 | |
4ETT
 
 | |
1GZ4
 
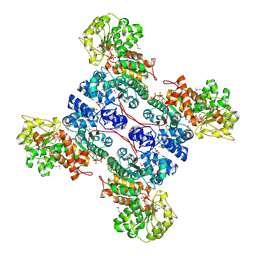 | | molecular mechanism of the regulation of human mitochondrial NAD(P)+-dependent malic enzyme by ATP and fumarate | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FUMARIC ACID, MANGANESE (II) ION, ... | | Authors: | Yang, Z, Lanks, C.W, Tong, L. | | Deposit date: | 2002-05-15 | | Release date: | 2002-07-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular Mechanism for the Regulation of Human Mitochondrial Nad(P)(+)-Dependent Malic Enzyme by ATP and Fumarate
Structure, 10, 2002
|
|
3KJP
 
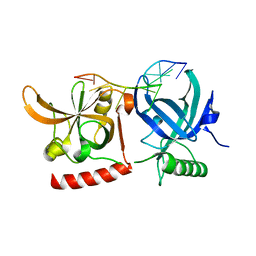 | | Crystal Structure of hPOT1V2-GGTTAGGGTTAG | | Descriptor: | 5'-D(*G*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*G)-3', Protection of telomeres protein 1 | | Authors: | Nandakumar, J, Cech, T.R, Podell, E.R. | | Deposit date: | 2009-11-03 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | How telomeric protein POT1 avoids RNA to achieve specificity for single-stranded DNA.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3BIK
 
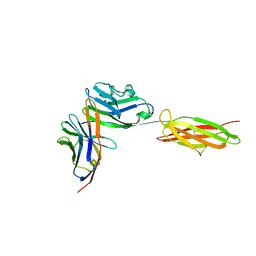 | | Crystal Structure of the PD-1/PD-L1 Complex | | Descriptor: | GLYCEROL, Programmed cell death 1 ligand 1, Programmed cell death protein 1 | | Authors: | Lin, D.Y, Tanaka, Y, Iwasaki, M, Gittis, A.G, Su, H.P, Mikami, B, Okazaki, T, Honjo, T, Minato, N, Garboczi, D.N. | | Deposit date: | 2007-11-30 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The PD-1/PD-L1 complex resembles the antigen-binding Fv domains of antibodies and T cell receptors.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3SMB
 
 | |
7TZC
 
 | | A drug and ATP binding site in type 1 ryanodine receptor | | Descriptor: | (2S)-3-(octadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 4-[(7-methoxy-2,3-dihydro-1,4-benzothiazepin-4(5H)-yl)methyl]benzoic acid, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Melville, Z, Dridi, H, Yuan, Q, Reiken, S, Anetta, W, Liu, Y, Clarke, O.B, Marks, A.R. | | Deposit date: | 2022-02-15 | | Release date: | 2022-05-18 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | A drug and ATP binding site in type 1 ryanodine receptor.
Structure, 30, 2022
|
|
3S5U
 
 | | Crystal structure of CRISPR associated protein | | Descriptor: | CALCIUM ION, Putative uncharacterized protein | | Authors: | Ke, A, Nam, K.H. | | Deposit date: | 2011-05-23 | | Release date: | 2011-06-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of clustered regularly interspaced short palindromic repeats (CRISPR)-associated Csn2 protein revealed Ca2+-dependent double-stranded DNA binding activity.
J. Biol. Chem., 286, 2011
|
|
4RH7
 
 | | Crystal structure of human cytoplasmic dynein 2 motor domain in complex with ADP.Vi | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ADP ORTHOVANADATE, ... | | Authors: | Schmidt, H, Zalyte, R, Urnavicius, L, Carter, A.P. | | Deposit date: | 2014-10-01 | | Release date: | 2014-12-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Structure of human cytoplasmic dynein-2 primed for its power stroke.
Nature, 518, 2015
|
|
4ZBT
 
 | | Streptomyces bingchenggensis aldolase-dehydratase in Schiff base complex with pyruvate | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Acetoacetate decarboxylase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mydy, L.S, Silvaggi, N.R. | | Deposit date: | 2015-04-15 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Sbi00515, a Protein of Unknown Function from Streptomyces bingchenggensis, Highlights the Functional Versatility of the Acetoacetate Decarboxylase Scaffold.
Biochemistry, 54, 2015
|
|
8R1G
 
 | | Dimeric ternary structure of E6AP-E6-p53 | | Descriptor: | Cellular tumor antigen p53, Ubiquitin-like protein SMT3,Protein E6, Ubiquitin-protein ligase E3A, ... | | Authors: | Sandate, C.R, Chakrabory, D, Kater, L, Kempf, G, Thoma, N.H. | | Deposit date: | 2023-11-01 | | Release date: | 2023-12-06 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Structural insights into viral hijacking of p53 by E6 and E6AP
Biorxiv, 2023
|
|
6RVW
 
 | | Structure of right-handed protein cage consisting of 24 eleven-membered ring proteins held together by gold (I) bridges. | | Descriptor: | GOLD ION, Transcription attenuation protein MtrB | | Authors: | Malay, A.D, Miyazaki, N, Biela, A.P, Iwasaki, K, Heddle, J.G. | | Deposit date: | 2019-06-03 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | An ultra-stable gold-coordinated protein cage displaying reversible assembly.
Nature, 569, 2019
|
|
6Z63
 
 | | FtsE structure from Streptococus pneumoniae in complex with ADP at 1.57 A resolution (spacegroup P 21) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division ATP-binding protein FtsE | | Authors: | Alcorlo, M, Straume, D, Havarstein, L.S, Hermoso, J.A. | | Deposit date: | 2020-05-27 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural Characterization of the Essential Cell Division Protein FtsE and Its Interaction with FtsX in Streptococcus pneumoniae.
Mbio, 11, 2020
|
|
5DCH
 
 | | Crystal structure of Pseudomonas aeruginosa DsbA E82I in complex with MIPS-0000851 (3-[(2-METHYLBENZYL)SULFANYL]-4H-1,2,4-TRIAZOL-4-AMINE) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[(2-methylbenzyl)sulfanyl]-4H-1,2,4-triazol-4-amine, GLYCEROL, ... | | Authors: | McMahon, R.M, Martin, J.L. | | Deposit date: | 2015-08-24 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.447 Å) | | Cite: | Fragment library screening identifies hits that bind to the non-catalytic surface of Pseudomonas aeruginosa DsbA1.
PLoS ONE, 12, 2017
|
|
5JYX
 
 | |
1H0J
 
 | | Structural Basis of the Membrane-induced Cardiotoxin A3 Oligomerization | | Descriptor: | CARDIOTOXIN-3, DODECYL SULFATE | | Authors: | Forouhar, F, Huang, W.-N, Liu, J.-H, Chien, K.-Y, Wu, W.-G, Hsiao, C.-D. | | Deposit date: | 2002-06-20 | | Release date: | 2003-06-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Membrane-Induced Cardiotoxin A3 Oligomerization
J.Biol.Chem., 278, 2003
|
|
2EPN
 
 | | N-acetyl-B-D-glucosaminidase (GCNA) from Streptococcus gordonii | | Descriptor: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, N-acetyl-beta-D-glucosaminidase | | Authors: | Langley, D.B. | | Deposit date: | 2007-03-30 | | Release date: | 2008-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structure of N-acetyl-beta-D-glucosaminidase (GcnA) from the Endocarditis Pathogen Streptococcus gordonii and its Complex with the Mechanism-based Inhibitor NAG-thiazoline
J.Mol.Biol., 377, 2008
|
|
4DM8
 
 | | Crystal structure of RARb LBD in complex with 9cis retinoic acid | | Descriptor: | Nuclear receptor coactivator 1, RETINOIC ACID, Retinoic acid receptor beta | | Authors: | Osz, J, Br livet, Y, Peluso-Iltis, C, Cura, V, Eiler, S, Ruff, M, Bourguet, W, Rochel, N, Moras, D. | | Deposit date: | 2012-02-07 | | Release date: | 2012-03-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for a molecular allosteric control mechanism of cofactor binding to nuclear receptors.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3SS3
 
 | |
6Z67
 
 | | FtsE structure of Streptococcus pneumoniae in complex with AMPPNP at 2.4 A resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division ATP-binding protein FtsE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Alcorlo, M, Straume, D, Havarstein, L.S, Hermoso, j.A. | | Deposit date: | 2020-05-28 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Characterization of the Essential Cell Division Protein FtsE and Its Interaction with FtsX in Streptococcus pneumoniae.
Mbio, 11, 2020
|
|
5DHZ
 
 | | HIV-1 Rev NTD dimers with variable crossing angles | | Descriptor: | Anti-Rev Antibody Fab single-chain variable fragment, heavy chain, light chain, ... | | Authors: | DiMattia, M.A, Watts, N.R, Wingfield, P.T, Grimes, J.M, Stuart, D.I, Steven, A.C. | | Deposit date: | 2015-08-31 | | Release date: | 2016-06-29 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | The Structure of HIV-1 Rev Filaments Suggests a Bilateral Model for Rev-RRE Assembly.
Structure, 24, 2016
|
|
1HFH
 
 | | SOLUTION STRUCTURE OF A PAIR OF COMPLEMENT MODULES BY NUCLEAR MAGNETIC RESONANCE | | Descriptor: | FACTOR H, 15TH AND 16TH C-MODULE PAIR | | Authors: | Barlow, P.N, Steinkasserer, A, Norman, D.G, Kieffer, B, Wiles, A.P, Sim, R.B, Campbell, I.D. | | Deposit date: | 1993-02-23 | | Release date: | 1993-07-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a pair of complement modules by nuclear magnetic resonance.
J.Mol.Biol., 232, 1993
|
|
2EPL
 
 | | N-acetyl-B-D-glucosaminidase (GCNA) from Streptococcus gordonii | | Descriptor: | GLYCEROL, N-acetyl-beta-D-glucosaminidase, SULFATE ION | | Authors: | Langley, D.B, Harty, D.W.S, Guss, J.M, Collyer, C.A. | | Deposit date: | 2007-03-30 | | Release date: | 2008-03-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of N-acetyl-beta-D-glucosaminidase (GcnA) from the Endocarditis Pathogen Streptococcus gordonii and its Complex with the Mechanism-based Inhibitor NAG-thiazoline
J.Mol.Biol., 377, 2008
|
|
