1BNN
 
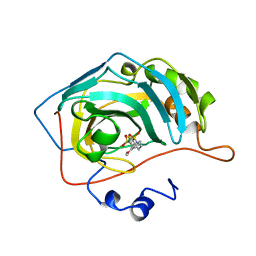 | | CARBONIC ANHYDRASE II INHIBITOR | | Descriptor: | 3,,4-DIHYDRO-2-(3-METHOXYPHENYL)-2H-THIENO-[3,2-E]-1,2-THIAZINE-6-SULFONAMIDE-1,1-DIOXIDE, CARBONIC ANHYDRASE, MERCURY (II) ION, ... | | Authors: | Boriack-Sjodin, P.A, Zeitlin, S, Christianson, D.W. | | Deposit date: | 1998-07-30 | | Release date: | 1999-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of inhibitor binding to human carbonic anhydrase II.
Protein Sci., 7, 1998
|
|
1BN4
 
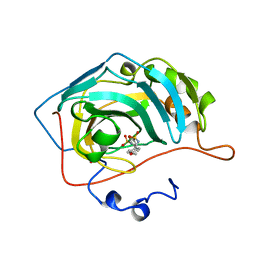 | | CARBONIC ANHYDRASE II INHIBITOR | | Descriptor: | CARBONIC ANHYDRASE, MERCURY (II) ION, N-[(4-METHOXYPHENYL)METHYL]2,5-THIOPHENEDESULFONAMIDE, ... | | Authors: | Boriack-Sjodin, P.A, Zeitlin, S, Christianson, D.W. | | Deposit date: | 1998-07-31 | | Release date: | 1999-05-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis of inhibitor binding to human carbonic anhydrase II.
Protein Sci., 7, 1998
|
|
1BN1
 
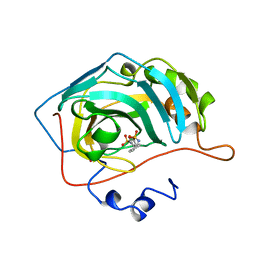 | | CARBONIC ANHYDRASE II INHIBITOR | | Descriptor: | CARBONIC ANHYDRASE, MERCURY (II) ION, THIOPHENE-2,5-DISULFONIC ACID 2-AMIDE-5-(4-METHYL-BENZYLAMIDE), ... | | Authors: | Boriack-Sjodin, P.A, Zeitlin, S, Christianson, D.W. | | Deposit date: | 1998-07-31 | | Release date: | 1999-05-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis of inhibitor binding to human carbonic anhydrase II.
Protein Sci., 7, 1998
|
|
1BNV
 
 | | CARBONIC ANHYDRASE II INHIBITOR | | Descriptor: | (S)-3,4-DIHYDRO-2-(3-METHOXYPHENYL)-4-METHYLAMINO-2H-THIENO[3,2-E]-1,2-THIAZINE-6-SULFONAMIDE-1,1-DIOXIDE, CARBONIC ANHYDRASE, MERCURY (II) ION, ... | | Authors: | Boriack-Sjodin, P.A, Zeitlin, S, Christianson, D.W. | | Deposit date: | 1998-07-30 | | Release date: | 1999-02-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analysis of inhibitor binding to human carbonic anhydrase II.
Protein Sci., 7, 1998
|
|
3EB2
 
 | |
3DZB
 
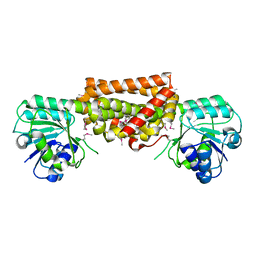 | |
3DZ1
 
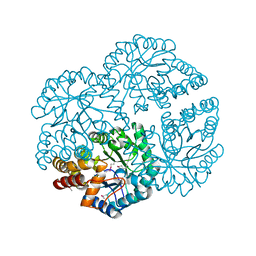 | |
3E82
 
 | | Crystal structure of a putative oxidoreductase from Klebsiella pneumoniae | | Descriptor: | CHLORIDE ION, Putative oxidoreductase | | Authors: | Eswaramoorthy, S, Mohammad, M.B, Thomas, C.A, Brown, A.C, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-08-19 | | Release date: | 2008-08-26 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of a putative oxidoreductase from Klebsiella pneumoniae
To be Published
|
|
3NQS
 
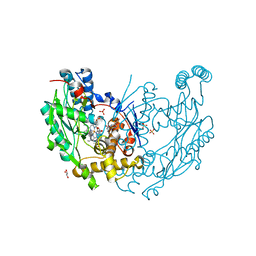 | | Crystal Structure of Inducible Nitric Oxide Synthase with N-Nitrosated-pterin | | Descriptor: | 1,2-ETHANEDIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, ETHYL 4-[(4-METHYLPYRIDIN-2-YL)AMINO]PIPERIDINE-1-CARBOXYLATE, ... | | Authors: | Rosenfeld, R.J, Getzoff, E.D, Tainer, J.A. | | Deposit date: | 2010-06-29 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Nitric-oxide synthase forms N-NO-pterin and S-NO-cys: implications for activity, allostery, and regulation.
J.Biol.Chem., 285, 2010
|
|
2CEN
 
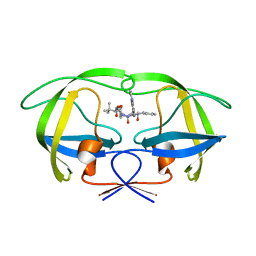 | | P1' Extended HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol in the Transition-State Mimicking Scaffold | | Descriptor: | POL PROTEIN, {(1S)-1-[N'-[(2S)-2-HYDROXY-2-((1S,2R)-2-HYDROXY-INDAN-1-YLCARBAMOYL)-3-PHENYL-PROPYL]-N'-[4-(PYRIDINE-2-YL)-BENZYL]-HYDRAZINOCARBONYL]-2,2-DIMETHYL-PROPYL}-CARBAMIC ACID METHYL ESTER | | Authors: | Ginman, N, Ekegren, J.K, Johansson, A, Wallberg, H, Larhed, M, Samuelsson, B, Hallberg, A, Unge, T. | | Deposit date: | 2006-02-08 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Microwave-Accelerated Synthesis of P1'-Extended HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol in the Transition-State Mimicking Scaffold.
J.Med.Chem., 49, 2006
|
|
2CEM
 
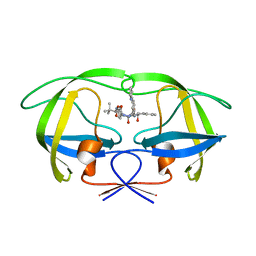 | | P1' Extended HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol in the Transition-State Mimicking Scaffold | | Descriptor: | POL PROTEIN, {(1S)-1-[N'-[(2S)-2-HYDROXY-2-((1S,2R)-2-HYDROXY-INDAN-1-YLCARBAMOYL)-3-PHENYL-PROPYL]-N'-[4-(PYRIDINE-2-YL)-BENZYL]-HYDRAZINOCARBONYL]-2,2-DIMETHYL-PROPYL}-CARBAMIC ACID METHYL ESTER | | Authors: | Ginman, N, Ekegren, J.K, Johansson, A, Wallberg, H, Larhed, M, Samuelsson, B, Hallberg, A, Unge, T. | | Deposit date: | 2006-02-08 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Microwave-Accelerated Synthesis of P1'-Extended HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol in the Transition-State Mimicking Scaffold.
J.Med.Chem., 49, 2006
|
|
2HAP
 
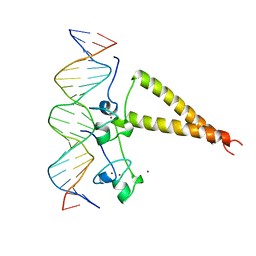 | | STRUCTURE OF A HAP1-18/DNA COMPLEX REVEALS THAT PROTEIN/DNA INTERACTIONS CAN HAVE DIRECT ALLOSTERIC EFFECTS ON TRANSCRIPTIONAL ACTIVATION | | Descriptor: | DNA (5'-D(*AP*CP*GP*CP*TP*AP*TP*TP*AP*TP*CP*GP*CP*TP*AP*TP*TP*AP*GP*T)-3'), DNA (5'-D(*AP*CP*TP*AP*AP*TP*AP*GP*CP*GP*AP*TP*AP*AP*TP*AP*GP*CP*GP*T)-3'), PROTEIN (HEME ACTIVATOR PROTEIN), ... | | Authors: | King, D.A, Zhang, L, Guarente, L, Marmorstein, R. | | Deposit date: | 1998-09-17 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of HAP1-18-DNA implicates direct allosteric effect of protein-DNA interactions on transcriptional activation.
Nat.Struct.Biol., 6, 1999
|
|
2VTK
 
 | |
1VTK
 
 | |
5M42
 
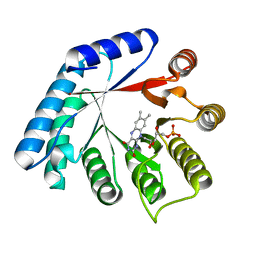 | | Structure of Thermus thermophilus L-proline dehydrogenase lacking alpha helices A, B and C | | Descriptor: | FLAVIN MONONUCLEOTIDE, Proline dehydrogenase | | Authors: | Martinez-Julvez, M, Huijbers, M.M.E, van Berkel, W.J.H, Medina, M. | | Deposit date: | 2016-10-18 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Proline dehydrogenase from Thermus thermophilus does not discriminate between FAD and FMN as cofactor.
Sci Rep, 7, 2017
|
|
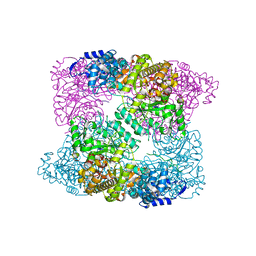
2XAP
 
 | |
6SLF
 
 | |
8C6Z
 
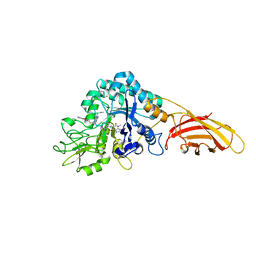 | | necrotic enteritis associated Clostridium p. chitinase F8UNI4 in complex with inhibitor bisdionin C | | Descriptor: | 1,1'-PROPANE-1,3-DIYLBIS(3,7-DIMETHYL-3,7-DIHYDRO-1H-PURINE-2,6-DIONE), CADMIUM ION, COBALT (II) ION, ... | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2023-01-12 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Contribution and structural characterization of Chitinases to Clostridium perfringens Pathogenesis in Broilers
To Be Published
|
|
3GPK
 
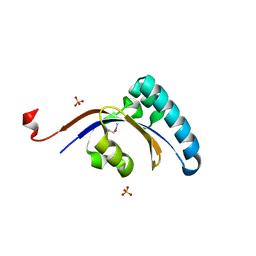 | |
7OWB
 
 | | Chimeric carminomycin-4-O-methyltransferase (DnrK) with a region from 10-hydroxylase CalMB | | Descriptor: | Carminomycin 4-O-methyltransferase DnrK,Methyltransferase domain-containing protein, S-ADENOSYL-L-HOMOCYSTEINE, methyl (1R,2R,4S)-2-ethyl-2,5,7-trihydroxy-6,11-dioxo-4-{[2,3,6-trideoxy-3-(dimethylamino)-alpha-L-lyxo-hexopyranosyl]oxy}-1,2,3,4,6,11-hexahydrotetracene-1-carboxylate | | Authors: | Dinis, P, MetsaKetela, M. | | Deposit date: | 2021-06-17 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Evolution-inspired engineering of anthracycline methyltransferases.
Pnas Nexus, 2, 2023
|
|
8OVR
 
 | |
3I3Y
 
 | |
4LE0
 
 | | Crystal structure of the receiver domain of DesR in complex with beryllofluoride and magnesium | | Descriptor: | ACETATE ION, BERYLLIUM TRIFLUORIDE ION, GLYCEROL, ... | | Authors: | Trajtenberg, F, Larrieux, N, Buschiazzo, A. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Allosteric activation of bacterial response regulators: the role of the cognate histidine kinase beyond phosphorylation.
MBio, 5, 2014
|
|
4LDZ
 
 | | Crystal structure of the full-length response regulator DesR in the active state | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Trajtenberg, F, Larrieux, N, Buschiazzo, A. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Allosteric activation of bacterial response regulators: the role of the cognate histidine kinase beyond phosphorylation.
MBio, 5, 2014
|
|
4Y1S
 
 | | Structural basis for Ca2+-mediated interaction of the perforin C2 domain with lipid membranes | | Descriptor: | CALCIUM ION, Perforin-1 | | Authors: | Conroy, P.J, Yagi, H, Whisstock, J.C, Norton, R.S. | | Deposit date: | 2015-02-09 | | Release date: | 2015-09-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.611 Å) | | Cite: | Structural Basis for Ca2+-mediated Interaction of the Perforin C2 Domain with Lipid Membranes.
J.Biol.Chem., 290, 2015
|
|
